
Identification of Ulva sp. Grown in Multitrophic Aquaculture Systems
*Corresponding Author(s):
Glauco FavotCollege Of Fisheries And Life Science, Shanghai Ocean University, Shanghai, China
Tel:+86 15601702825,
Email:a53585@ualg.pt
Abstract
The genus Ulva is one of the most numerous of marine and estuarine genera. Traditionally cultivated for human consumption, since the 1990s Ulva was integrated into land based Integrated Multi-Trophic Aquacultures (IMTA) for biomass production and bioremediation. A proper taxonomic identification is the first critical step in implementing an algal production program. However, Ulva species are difficult to morphological identify due to their phenotypic plasticity. Combined molecular and morphological techniques can lead to better characterization of Ulva sp. This study identifies the Ulva sp. cultivated in earth ponds facing the Ria Formosa Lagoon (South Portugal) as well as green algae growing spontaneously in the ponds. DNA barcoding with the markers ITS (Internal Transcribed Spacer) identified six species, with Ulva flexuosa being the cultivated one. Ulva flexuosa was recorded for the first time in South Portugal. However, taxonomic questions were raised because distinct clades were found for this species using published sequences. The ‘lettuce-leaf’ morphotype observed is not attributable to any of the marine subspecies of Ulva flexuosa.
Keywords
DNA-Barcoding; ITS; Species identification; Ulva flexuosa
INTRODUCTION
The genus Ulva is one of the most numerous of marine and estuarine genera [1]. The cosmopolitan distribution of the genus Ulva makes it suitable for cultivation practically everywhere [2]. Traditionally cultivated for human consumption, since the 1990s Ulva was integrated into land based Integrated Multi-Trophic Aquacultures (IMTA) for biomass production and bioremediation [3]. Ulva spp. withstand the extreme environmental condition of earth ponds and when grown in effluent media, protein content increases (> 40%), resulting in a valuable feed for macroalgivore species with high commercial value [2-5]. The current market for these algae is limited, but could see growth considering the suitability of Ulva as a biomass energy resource and its application as a raw material for nutraceuticals, biomaterials and sulphated polysaccharides (Ulvan) [3,5-7]). Given the growing demand for algae, a proper taxonomic identification is necessary in aquaculture [8,9]. Selecting appropriate target species is the critical first step in implementing an algal production programme. Moreover, improper taxonomic identification makes comparing results difficult, inhibiting the consolidation of knowledge about production and other characteristics of cultivated species [9]. An accurate assessment of marine macroalgae is important for conservation, monitoring, and management of biological introductions and invasions [10]. Due to phenotypic plasticity, the morphological characteristics of several Ulva species have insufficient taxonomic value [11-13]). The combination of molecular and morphological techniques can lead to better characterization of taxa [14-18]). DNA barcoding is a taxonomic method that uses a short genetic marker in an organism’s DNA to identify it [19]. The main goal is to identify an unknown sample in terms of a pre-existing classification [20]. The Internal Transcribed Spacer region of ribosomal cistron (ITS) has been used in several studies concerning the Ulva species identification [16,21,22]). ITS is proving useful for identification at species level due to its multiple highly variable regions [23-25].
The IPMA aquaculture research station in Olhão (EPPO-EstaçãoPiloto de Piscicultura de Olhão), Portugal, cultivated Ulva sp. during an Integrated Multitrophic Aquaculture (IMTA) experiment in earthen ponds. The purpose of this study was to verify the taxonomic identity of the Ulva sp. grown and identify the green algae that grew spontaneously in the pond system.
MATERIALS AND METHODS
The IMTA experiment was conducted at the Aquaculture Research Station in Olhão (EPPO-Estação Piloto de Piscicultura de Olhão), Portugal. The EPPO station is located in the salt marshes of Ria Formosa coastal lagoon, a mesotidal system in the south of Portugal (Figure 1a). Ulva spp. were collected in the main discharge channel of EPPO (Figure 1b), a portion was weighted and individually planted in 6 rafts (1 m2 each), made of horizontal nets stretched between styrofoam floaters (Figure 2a).
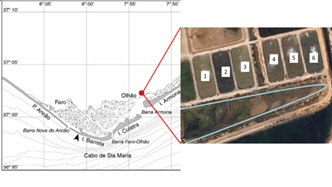 Figure 1: a) Schematic representation of the Ria Formosa lagoon system [26]; b) Satellite image of the ponds used for the IMTA experiment (indicated by numbers); in light blue the discharge channel from which Ulva sp. was collected.
Figure 1: a) Schematic representation of the Ria Formosa lagoon system [26]; b) Satellite image of the ponds used for the IMTA experiment (indicated by numbers); in light blue the discharge channel from which Ulva sp. was collected.
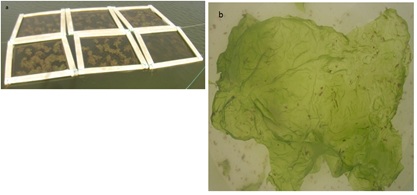 Figure 2: a) The six floating rafts structure used to cultivate b) Ulva sp.
Figure 2: a) The six floating rafts structure used to cultivate b) Ulva sp.
Collection and storage of seaweeds
In the beginning of November (3/11/2016), 54 samples of green seaweeds were collected from the earthen ponds, including 17 from the floating rafts. The remaining samples were collected on the perimeter of the ponds or rafts (e.g. ropes). Subsequently, each sample was washed clean with seawater and thoroughly dried with absorbent paper. Of each specimen, a piece of approximately 1 cm2 was preserved in silica. Each bag was labeled with the date of withdrawal, the pond number, letter “f” or “t” (Framework or Pond), and sample number. The remainder of each individual collected was preserved as herbarium voucher. This identification system allowed a visual comparison after the species were identified through Barcoding.
DNA extraction.
Silica dried algal biomass was prepared for the DNA extraction through homogenizing the samples by grinding with a tungsten sphere in a mixer mill (Eppendorf A-2-DWP) for 3 minutes at max speed (3,700 rpm). Seaweed DNA was extracted using the NucleoSpin® Plant II Kit (MACHEREY-NAGEL GmbH & Co. KG, Germany) following the manufacture’s protocol.
The quality of the DNA was verified by running 5 µl of the DNA extraction (with 1 µl Gel-Red and 2 µl of loading buffer (5 X Green GoTaq Flexi Buffer)) of six randomly selected samples on a 0.8 % agarose gel.
DNA amplification and sequencing.
The nuclear primers ITS1 5’-TCCGTAGGTGAACCTGCGG-3’ and ITS4 3’-CGTATAGTTATTCGCCTCCT-5’ were used to amplify nuclear rDNA (Ribosomal DNA) fragment [27]. This fragment contains the Internal Transcribed Spacer 1 (ITS1), the 5.8 S gene, and the Internal Transcribed Spacer 2 (ITS2) [27]. Each pcr reaction consisted of 23.95 µl H2O Milli-Q, 4 µl of 5 X Buffer, 1.6 µl 25 mM Mg, 1.25 µl 2 mM of each dNTP, 2 µl 1.0 µM of each primer, 0,2 µl 5 U/µl Go-Taq, 5.0 µL of diluted (1:100 H2O Milli-Q) genomic DNA extract.
PCR amplification was run a Applied Biosystems 2720 Thermal Cycler (Applied Biosystems™, Foster City, CA) and the pro?le of the reaction consisted of an initial denature at 95°C for 5 min followed by 35 cycles of 95°C for 30s, 55°C for 30s min and 72°C for 1 min, and a ?nal extension at 72°C for 10 min.
The 54 PCR products were visually checked on a stained electrophoreses gel (2 % agarose). PCR products consisting of a single band with the right size were sequenced. DNA sequencing was performed on an ABI 3130 × l capillary sequencer (Applied Biosystems - CCMAR, Portugal) using the forward primers that were used for PCR.
Molecular analysis
The generated sequences were trimmed and aligned manually using Geneious R7.1.9 [28]. Subsequently identification was based on their DNA sequences by comparing them with sequences present in Genbank [29]. This operation was performed using Nucleotide BLAST web interface [30].
Phylogenetic analyses - alignment
DNA sequence alignment was created using the best quality sequence of each Ulva recognized in this study and from respective sequences chosen from BLAST results. Additional sequences for phylogenetic calculation were downloaded from Genbank selecting from other species used in previous papers [11,16,18,22] (Annex, Table 1).
|
Taxa |
Collection sites |
Source |
Accession Number ITS |
|
Ulvaria obscura spp. Blytii ((Areschoug) Bliding, 1969) |
Padilla Bay, WA, USA |
Hayden et al. [45] |
AY260571 |
|
Ulva californica (Wille in Collins, Holden et Setchell, 1899) |
La Jolla, CA, USA |
Hayden et al. [45] |
AY260560 |
|
Ulva californica (Wille in Collins, Holden et Setchell, 1899) |
Northeast Pacific |
Lawton et al. [18] |
AY422515 |
|
Ulva clathrata ((Roth) C. Agardh, 1811) |
Yellow Sea, China |
Teng et al. 2010 |
HQ197901 |
|
Ulva flexuosa (Wulfen,1803) |
Oshoro, Hokkaido, Japan |
Shimada et al. [11] Lawton et al. [18] |
AB097644 |
|
Ulva flexuosa spp. pilifera (Kützing), M.J.Wynne 2005 |
Poland |
Mareš et al. [16] Rybak et al. [22] |
HM447579 |
|
Ulva flexuosa spp. paradoxa (( C.Agardh) M.J.Wynne, 2005) |
Czech Republic |
Mareš et al. [16] Rybak et al. [22] |
HM447561 |
|
Ulva flexuosa spp. flexuosa (Wulfen, 1803) |
Sweden |
Mareš et al. [16] Rybak et al. [22] |
HM447564 |
|
Ulva lactuca (Linneus, 1753) |
N.A.* |
Mareš et al. [16] Rybak et al. [22] |
AJ234310 |
|
Ulva lactuca (Linneus, 1753) |
Northeast Pacific |
Mareš et al. [16] Rybak et al. [22] |
AY422499 |
|
Ulva linza (Linneus, 1753) |
Humboldt Bay, CA, USA |
Hayden et al. [45] |
AY260557 |
|
Ulva procera (K.Ahlner) Hayde, et al. [45] |
N.A. |
Hayden et al. [45] |
AY260558 |
|
Ulva procera |
Northeast Pacific |
Mareš et al. [16] Rybak et al. [22] |
AY422521 |
|
Ulva prolifera |
Yellow Sea (China) |
Zhang [57] |
KT802960 |
|
Ulva pseudocurvata (Koeman et van den Hoek, 1981) |
N.A. |
Mareš et al. [16] Rybak et al. [22] |
AJ234312 |
|
Ulva rigida |
Northeast Pacific |
Mareš et al. [16] Rybak et al. [22] |
AY422522 |
|
Ulva sapora |
Shelly Beach, Caloundra Australia |
Phillips et al. [35] |
KT374006 |
|
Ulva scandinavica |
N.A. |
Mareš et al. [16] Rybak et al. [22] |
AJ234317 |
|
Ulva taeniata ((Setchell) Setchell et Gardner, 1920) |
Monterey, CA, USA |
Mareš et al. [16] Rybak et al. [22] |
AY422525 |
|
Ulva tanneri |
Northeast Pacific |
Mareš et al. [16] Rybak et al. [22] |
AY422519 |
|
Ulva torta |
Fukui (Japan) |
Ogawa et al. [58] |
AB830503 |
|
Ulva torta |
Clovelly, NSW (Australia) |
Lawton et al. [18] |
KF195491 |
|
*N.A.: Not available |
|||
Table 1: Sources of taxa used to create the phylogenetic trees.
Initial alignment of the nucleotide sets was obtained using Geneious R7.1.9 [28]. Subsequently, the sequences were trimmed to a standard length and the identical sequences removed. The final alignment contained 33 taxa (32 in group taxa plus one outgroup (Ulvaria obscura)), of which five sequences from this study. The alignment was realigned with MAFFT v. 7.310 online applications using Q-INS-I algorithm (with default parameters) [31]. The lasts adjustments of the resulting alignments were carried out using Geneious.
Phylogenetic analyses - construction of phylogenetic tree
The phylogenetic analyses were performed using the Maximum-Likelihood (ML) and Bayesian Inference (BI) methods [32]. The ML tree was obtained using the PhyML online program [33] and the BI tree was constructed using MrBayes present in Geneious R7.1.9. The program jModelTest version 2.1.10 [34] was used to find the model of sequence evolution that best fit the dataset. ML and Bayesian trees were built using the Generalized Time Reversible (GTR) substitution model with discrete gamma distribution in four categories. One thousand bootstrap replications were performed for both methods using default setting to compare relative support of branches.
The phylogenetic analyses, nucleotide homology (%) and sequence divergence (bp) estimates were based on 520 bp, including gaps (Annex, Table 2).
|
Clade |
Species |
Collection sites |
Accession Number ITS |
Homology % |
D.B.S (bp)* |
|
A |
Ulva flexuosa T11t4 |
EPPO pond |
|||
|
Ulva flexuosa |
Oshoro, Hokkaido, (Japan) |
AB097644 |
99.47 |
2 |
|
|
Ulva californica |
La Jolla, California (U.S.A.) |
AY260560 |
97.33 |
12 |
|
|
Ulva californica |
Northeast Pacific |
AY422515 |
96.8 |
14 |
|
|
B |
Ulva torta T16t2 |
EPPO pond |
|||
|
Ulva torta |
Fukui (Japan) |
AB830503 |
95.65 |
17 |
|
|
Ulva torta |
Clovelly, NSW (Australia) |
KF195491 |
94.39 |
20 |
|
|
Ulva clathrata T15t6 |
EPPO pond |
95.17 |
19 |
||
|
Ulva clathrata |
Yellow Sea (China) |
HQ197901 |
94.91 |
22 |
|
|
B |
Ulva clathrata T15t6 |
EPPO pond |
|||
|
Ulva clathrata |
Yellow Sea, (China) |
HQ197901 |
99.49 |
2 |
|
|
Ulva torta |
Fukui (Japan) |
AB830503 |
97.71 |
9 |
|
|
Ulva torta |
Clovelly, NSW (Australia) |
KF195491 |
95.69 |
17 |
|
|
Ulva torta T16t2 |
EPPO pond pond |
95.17 |
19 |
||
|
C |
Ulva prolifera |
EPPO pond |
|||
|
Ulva prolifera |
Yellow Sea (China) |
KT802960 |
98.6 |
5 |
|
|
D |
Ulva intestinalis |
EPPO pond |
|||
|
Ulva sapora |
Shelly Beach, Caloundra (Australia) |
KT374006 |
96.48 |
14 |
|
|
*Distance between sequences (base-pair) |
|||||
Table 2: Nucleotide homology (%) of ITS region sequences of the EPPO samples and other Ulva specimens available in GenBank, that grouped in the ITS phylogenetic tree.
Analysis of morphology and anatomy
Morphology of thalli was assessed for fresh algae using a Nikon SMZ 1000 Stereo microscope whereas for anatomy a Nikon H550S Microscope (© 2017 Nikon Instruments Europe B.V) was used. All photos were captured and prepared using Nis-Elements Software (© 2017 Nikon Instruments Europe B.V).
RESULTS
Molecular analysis
Of the 54 samples used for molecular analysis 24 had the required high quality for analyses. The molecular analysis of the macroalgae collected from the EPPO ponds established that the Ulva cultivated in the rafts during the IMTA experiment was Ulva flexuosa (Wulfen, 1803). In addition, 5 other Ulva and 2 Cladophora species were identified from the pond system (Annex, Table 3).
The Ulva genus was well represented and consisted of: Ulva flexuosa (Wulfen, 1803, xxii,1), Ulva clathrata ((Roth) C. Agardh, 1811: 23), Ulva intestinalis (Linnaeus, 1753: 1163), Ulva sapora[1] [35], Ulva torta ((Mertens) Trevisan, 1842: 480) and Ulva prolifera (O.F.Müller, 1778: 7). Ulva sapora sequence obtained had a bad quality (5.5%) and was omitted from the phylogenetic analysis.
Phylogenetic trees
The phylogenetic analyses performed with the ML (Maximum Likelihood) and BI (Bayesian Inference) methods gave comparable tree topologies with the Ulva species coming from the ponds forming four distinct groups (Figures 3 & 4). These four groups, well supported both in the ML and BI trees, consist of: Two monophyletic (C,D) groups, one polyphyletic (A) group and in group B) U.torta is paraphyletic with respect to U.clathrata. However, the internal nodes are well supported only in the BI tree, with Bayesian Inference Posterior probability (BP) between 56 % and 86 %.
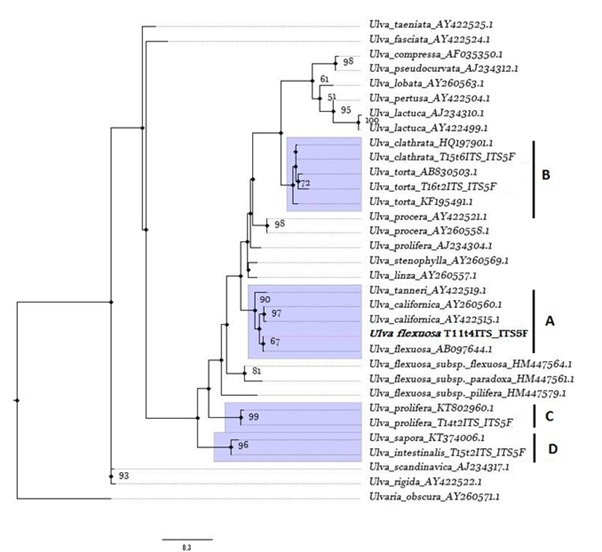 Figure 3: Maximum-Likelihood (ML) tree of ITS sequences calculated using the evolution model GTR+I+G. ML bootstrap values (1,000 replications) are given on the branches. Bootstrap support < 50 % are not shown. Sequences are labelled with taxon name and GenBank accession number of ITS sequence (Annex, Table 1). The tree is rooted using Ulvaria obscura; A, B, C and D refer to clades containing Ulva collected from EPPO ponds. In bold is stressed the Ulva flexuosa identified in this study.
Figure 3: Maximum-Likelihood (ML) tree of ITS sequences calculated using the evolution model GTR+I+G. ML bootstrap values (1,000 replications) are given on the branches. Bootstrap support < 50 % are not shown. Sequences are labelled with taxon name and GenBank accession number of ITS sequence (Annex, Table 1). The tree is rooted using Ulvaria obscura; A, B, C and D refer to clades containing Ulva collected from EPPO ponds. In bold is stressed the Ulva flexuosa identified in this study.
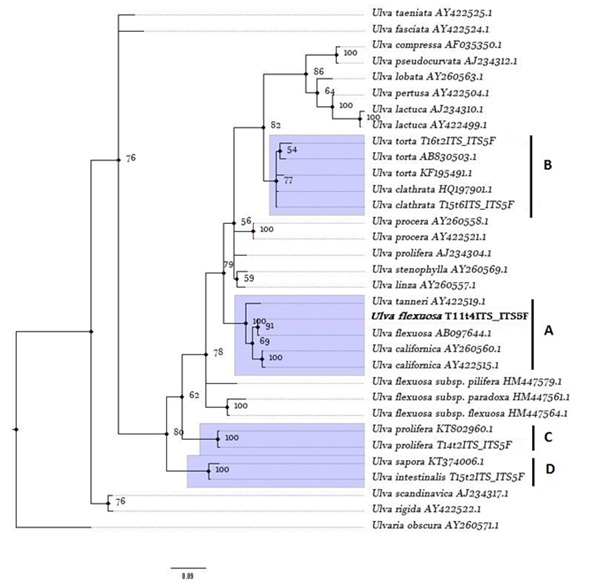 Figure 4: Bayesian tree of ITS sequences. Bayesian Probabilities (%), BP, are given on the branches. Posterior probabilities < 50 % have been omitted. Sequences are labelled with taxon name and GenBank accession number of ITS sequence (Annex, Table 1). The tree is rooted using Ulvaria obscura. A, B, C and D refer to class containing Ulva collected from EPPO ponds. In bold is stressed the Ulva flexuosa identified in this study.
Figure 4: Bayesian tree of ITS sequences. Bayesian Probabilities (%), BP, are given on the branches. Posterior probabilities < 50 % have been omitted. Sequences are labelled with taxon name and GenBank accession number of ITS sequence (Annex, Table 1). The tree is rooted using Ulvaria obscura. A, B, C and D refer to class containing Ulva collected from EPPO ponds. In bold is stressed the Ulva flexuosa identified in this study.
Group A showed that Ulva flexuosa present in the pond system formed a monophyletic clade with Ulva flexuosa from Hokkaido, Japan, with a nucleotide homology of 99.47 % (2 bp difference) (Table 1). According to this phylogram, either U.flexuosa are closely related to monophyletic group of Ulva californica (internal node value of 69 %) and the nucleotide homology showed between two species (≈ 97 %)supported a high similarity between these taxa. The Ulva flexuosa identified showed a low similarity with other European U.flexuosa subspecies with nucleotide homology < 87.9 % (Table 2).
|
|
U.flexuosa_ T11t4ITS |
U.flexuosa AB097644 |
U.californica AY260560 |
U.californica AY422515 |
|
U.flexuosa T11t4ITS |
- |
|
|
|
|
U.flexuosa AB097644 |
99.47 |
- |
|
|
|
U.californica AY260560 |
97.43 |
96,81 |
- |
|
|
U.californica AY422515 |
96.80 |
96,28 |
99,47 |
- |
Table 1: Nucleotide homology (in percentage) of ITS region sequences of the four species present in the clade of Ulva flexuosa grown within the ponds. In red is stressed the nucleotide homology of U.flexuosa grown in the ponds.
|
|
U. flexuosa_ T11t4ITS
|
U.flexuosa subsp. flexuosa HM447564 |
U.flexuosa subsp. paradoxa HM447561 |
U.flexuosa subsp. pilifera HM447579 |
|
U. flexuosa T11t4ITS |
_ |
|
|
|
|
U.flexuosa subsp. flexuosa HM447564 |
87.90 |
_ |
|
|
|
U.flexuosa subsp. paradoxa HM447561 |
84.30 |
91,71 |
_ |
|
|
U.flexuosa subsp. pilifera HM447579 |
85.75 |
90.52 |
85.53 |
_ |
Table 2: Nucleotide homology (in percentage) of ITS region sequences between Ulva flexuosa grown within the ponds and European Ulva flexuosa subspp. In red is stressed the nucleotide homology of U. flexuosa grown in the ponds.
Also the groups C, B and D were well supported and showed that all Ulva species sampled were closely related with the species from the North Pacific (nucleotide homology between ≈ 99 % to ≈ 96 %) (Annex, Table 2).
Morphological observations
The gross morphological characteristics (Annex, Table 3) presented a marked homogeneity among the varied species collected, underlining the importance of genetic analysis to identify the different Ulva species. The filamentous, herbaceous-like shape was the most common and, with a few exceptions of turf forms (Ulva sapora and one Ulva clathrata), Ulva flexuosa was the only species present with 3 different dominant morphotypes:
- The lettuce-leaf (Figure 5a-5b).
- Narrow and broad gregarious thalli (Figure 5c).
- Filamentous, herbaceous-like shape (Figure 6a-e).
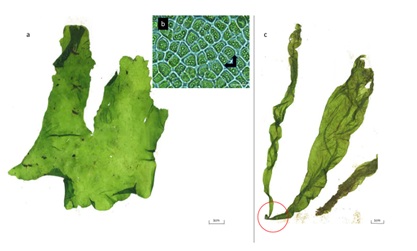 Figure 5: a) Lettuce-shape Ulva flexuosa; b) polygonal cells with pyrenoids (black rows); c) Gregarious thalli with discoidal base (red circle). Scale bar a) and c) 1cm. Scale bar for b) is 10µm.
Figure 5: a) Lettuce-shape Ulva flexuosa; b) polygonal cells with pyrenoids (black rows); c) Gregarious thalli with discoidal base (red circle). Scale bar a) and c) 1cm. Scale bar for b) is 10µm.
 Figure 6: a) Ulva flexuosa filamentous morphotype; b) thallus corrugated; c) laminar; d) branch (red circle); e) hollow stipe. Scale bar a) 1cm; scale bars of b), c), d) and e) are 1mm.
Figure 6: a) Ulva flexuosa filamentous morphotype; b) thallus corrugated; c) laminar; d) branch (red circle); e) hollow stipe. Scale bar a) 1cm; scale bars of b), c), d) and e) are 1mm.
|
Sample |
Description |
Morphological assessment |
|
11-t3 |
Ulva flexuosa (Wulfen,1803) |
Filamentous, herbaceous shape |
|
11-t4 |
Ulva flexuosa (Wulfen,1803) |
Filamentous, herbaceous shape |
|
11-f2 |
Ulva flexuosa (Wulfen,1803) |
Lettuce-leaf, flat, rounded undulate margins. |
|
16-t1 |
Ulva flexuosa (Wulfen,1803) |
Lettuce-leaf, flat, rounded undulate margins. |
|
16-t2 |
Ulva torta ((Mertens) Trevisan, 1842) |
Narrow small leaf, rounded on top. |
|
16-t5 |
Ulva flexuosa (Wulfen,1803) |
Linear compress thalli, tapering toward the base. |
|
16-t6 |
Ulva sapora (Phillips et al. [35])* |
Turf form, Thin-short filamentous |
|
16-f3 |
Ulva flexuosa (Wulfen,1803) |
Filamentous, tubular and linziformis. |
|
16-f4 |
Ulva flexuosa (Wulfen,1803) |
Filamentous, herbaceous shape |
|
12-t2 |
Cladophora albida ((Nees) Kutzing, 1843) |
Dark green, musk form |
|
12-t3 |
Cladophora vagabunda ((Linnaeus) Hoek, 1963) |
Narrow liner flat leaf |
|
12-t5 |
Ulva flexuosa (Wulfen,1803) |
Filamentous, herbaceous shape |
|
14-t2 |
Ulva prolifera (O.F.Müller, 1778) |
Filamentous, herbaceous shape |
|
14-t3 |
Ulva flexuosa (Wulfen,1803) |
Filamentous, herbaceous shape |
|
13-t2 |
Ulva flexuosa (Wulfen,1803) |
Linear compress thalli, herbaceous shape. |
|
13-t5 |
Ulva flexuosa (Wulfen,1803) |
Lettuce-Leaf, flat, rounded edges, undulate margin |
|
13-t6 |
Ulva flexuosa (Wulfen,1803) |
Lanceolate Leaf. |
|
13-t8 |
Ulva clathrata ((Roth) C.Agardh, 1811) |
Filamentous, herbaceous shape |
|
13-f2 |
Ulva flexuosa (Wulfen,1803) |
Filamentous, herbaceous shape |
|
13-f3 |
Ulva flexuosa (Wulfen,1803) |
Lettuce-leaf present some perforation |
|
15-t2 |
Ulva intestinalis (Linnaeus, 1753) |
Tubular, herbaceous shape |
|
15-t3 |
Ulva flexuosa (Wulfen,1803) |
Narrow and broad gregarious thalli, small discoid base |
|
15-t4 |
Ulva flexuosa (Wulfen,1803) |
Linear compress thalli, round on top. |
|
15-t6 |
Ulva clathrata ((Roth) C.Agardh, 1811) |
Turf form, Thin-short filamentus |
|
*This name is currently regarded as a synonym of Ulva tepida (Masakiyo and S. Shimada, 2014) (Algaedatabased). |
||
Table 3: Ulva taxa identified with short morphological description.
The lettuce-like Ulva flexuosa was the cultivated one. The specimen had a less rigid structure (thin and papery in texture) than those collected in the drainage channel. Moreover, they lost any anchoring structure present in the wild type. Their thalli had medium to light green, broader than long, flat, irregular contoured with undulated margins and was unbranched (Figure 5a). Under the microscope the central part of lettuce-like’s thallus showed a disordered cell arrangement with 2-4 pyrenoids per cell. Cells were irregularly arranged, polygonal, usually with rounded corners (Figure 5b). Principally measurements are shown in table 3.
|
U.flexuosa |
Length of cells (µm) |
Width of cells(µm) |
ø of pyrenoids |
Nº of pyrenoids (in one cells) |
|
Mean |
8.04 |
5.61 |
1.84 |
3.5 |
|
Min. |
5.19 |
1.99 |
0.97 |
1 |
|
Max. |
11.27 |
5.87 |
2.91 |
4 |
|
SD* |
1.2 |
1.08 |
0.42 |
|
|
*SD= Standard Deviation |
||||
Table 3: Size of Ulva flexuosa cells with wide leaf thalli.
The two remaining morphotypes belong to Ulva flexuosa grown within the ponds or attached to the framework. The first of these was characterized by a narrow and broad gregarious thallus attached to substrate by means of a small discoid base and like the cultivated morphotype was unbranched, flat with a thin texture and, started from a narrow base, widening towards the top. The U.flexuosa on the framework had a filamentous herbaceous shape and it often presented thalli polyform, slender, tubular compressed or laminar, widening at the top. Observations under the stereoscope revealed the presence of some branches at the base and a stipe that could be hollow. The thalli were fixed by means of a basal disc reinforced by numerous robust rhizoidal filaments. It is worth mentioning the presence of a fourth morphotype, with lanceolate thallus, although it is represented by a single specimen collected around the 13-pond’s perimeter.
[1]Ulva sapora is a synonymous name of Ulva tepida (Masakiyo, Y. & Shimada, S. (2014) [36]) discovered in Japan for the first time and then reported in Australia (Phillips et al. 2016 [35]) as not indigenous species.
DISCUSSION
The identification of Ulva spp. present in the EPPO ponds revealed a heterogeneous community, consisting of six taxa of which two were never reported in the Ria Formosa area: Ulva flexuosa and Ulva torta.
Ulva flexuosa was identified as the species cultivated and its lettuce-leaf morphotype is not attributable to any of the subspecies of this marine species.
The ITS allowed to differentiate Ulva taxa among our samples. The huge morphological plasticity of the kind probably would otherwise have led to associate the different phenotypes found with a species already recorded in the Ria Formosa. The presence of multiple bands in PCR products has already been reported in the past [24,37]. Therefore, ITS is commonly associated with rbcL (plastid rubisco large subunit) marker to increase the successes of identification [1,11,15,16,22,23,38].
Ulva flexuosa
U.flexuosa was originally described by Wulfen from the Adriatic Sea in the 1803. Currently, Ulva flexuosa species includes 4 subspecies and one variety: U.flexuosa ssp. flexuosa (Wulfen 1803: xxii, 1), U.flexuosa ssp. paradoxa (C. Agardh) M.J. Wynne [39], U.flexuosa var. linguiformis [13], U.flexuosa ssp. biflagellata (Bliding) A. Sfriso & D. Curiel [40] and U.flexuosa ssp. pilifera (Kützing) M.J. Wynne [39,41,42]. Among the three morphotype here reported the lettuce-leaf observed is not referable to any of the marine subspecies belonging to Ulva flexuosa. However, previous studies recorded a similar phenotype for the freshwater Ulva flexuosa ssp. pilifera [16,42]). Perhaps this morphotype may be explained considering algae grown in IMTA systems tend to develop leaves larger than wild types [43]. The remaining two morphologies have a taxonomic response. The filamentous one, based on the polymorphism of the thallus and the presence of a tubular stipe, could be associated to Ulva flexuosa ssp. flexuosa [13]. The gregarious thalli, instead, was similar to the Ulva flexuosa morphotype described by Wolf et al. [17] in the Venice lagoon, Ulva flexuosa from Busan and Pohang, Korea [44] and with the U.flexuosa ssp. pilifera identified in a recent study in the Polish freshwater [42]. However, genetic identity discarded the hypothesis of three distinct subspecies confirming instead the enormous plasticity of Ulva genus. There are several factors that can explain this phenomenon. Ulva flexuosa can ‘switch’ its thallus morphotype from tubular to foliose along their life and it is more frequent in culture due to stresses unique to artificial systems [22,45]. The place where the thalli develop (e.g. bottom or water surface) and environmental factors such as salinity and temperature can also affect morphological plasticity [25,42]). In our case, the fact of having collected seaweed in November after a week of intense rain may have favoured the finding of different morphotypes due to lowering of the temperature and salinity. Furthermore, in the past the role of bacterial community on morphology variation of Ulva genus has been shown [46,47]). The capacity of Crassostrea gigas to remove large amounts of bacteria [48] could perhaps have provoked a change in their community promoting change in Ulva flexuosa phenotype. All these possibilities need further studies.
Historically this species has been recorded in neighbouring countries along the coastal zone between Tanger (Morocco) and Melilla (Spain) [49] and in the Cadiz Bay [50]. Furthermore, U.flexuosa has been included in the list of macroalgae of the North coast of Portugal, along Minho, Douro Litoral, and Beira Litoral regions [51] and in Corunna harbour, Spain [52].
The Ulva flexuosa T11t4 sequence turned out to be almost identical (2 bp of difference) to that recorded by Shimada in Hokkaido, Japan [11] forming a well-defined clade in both phylogenetic trees. This observation may suggest the origin of these macroalgae could be the North Pacific and other studies suggest a common origin between the Ulva flexuosa of South Europe and the Pacific. An investigation about cryptic (species with morphologies identical or similar, although genetically different [17] and new species in the North Adriatic reported an Ulva flexuosa quite identical to one reported in British Columbia (Canada) [17]. Moreover, a Greek Ulva flexuosa var. linguiformis was closer related with a Japanese specimen [11,13,41]).
The Ulva flexuosa specimens from the EPPO ponds and South Europe did not match genetically with Ulva flexuosa subspecies from North Europe [16,22]), as was already detected by Marês and Shimada [16,23]. Marês et al. [16] proposed to indicate U.flexuosa as indigenous species of the inland waters of the Europe proposing a different nomenclature for the Asian, however, no mention was made about marine Ulva flexuosa.
Other taxa
Not only Ulva flexuosa was recorded for the first time in the Ria Formosa lagoon, also Ulva torta was first reported whereas Ulva intestinalis, Ulva prolifera and Ulva clathrata have been already mentioned in some studies that took place in the lagoon [53,54]. Historically, all these taxa, with sometimes the exception of Ulva torta, have shown a similar geographical distribution, jointly with U.flexuosa, in Portugal and neighboring countries [49-52]. Moreover, in the port of Corunna they occupied the same environment [52]. Nevertheless, among the studies listed above only Alsufyani et al. [54] provided a molecular identification by means of molecular techniques. This can lead to some doubts about the real distribution of this species.
CONCLUSION
The presence of Ulva flexuosa in the South Portugal broadens its geographic distribution in the country. The use of the molecular marker ITS was successful on macroalgae cultivated but there was low amplification success. For this reason subsequent investigations of green macroalgae would require the use of markers with a higher success rate such as tufA or associating rbcL (plastid rubisco large subunit) with the use of ITS [24]. Two studies in Poland and the USA leaded to the hypothesis that macroalgae previously identified as subspecies of Ulva flexuosa may be young species undergoing separation due to isolation and adaptation to different habitats [42,55]). Hence the recommendation to investigate into the phylogenetic relationships between U.flexuosa subspecies using more sensitive and specific molecular markers (e.g. ISSR (Inter-Simple Sequence Repeat), or SCAR (Sequence Characterized Amplified Region)) [42,56]). The genetic data collected in this experiment may lead to conclude that the origin of the macroalgae present in EPPO ponds could be the North Pacific. However, the scale of the present study does not allow to state which is the actual distribution area of the Ulva spp. identified and their status of native or introduced species. The presence of several species of Ulva suggests they withstand the ponds environment and proposes Ulva spp. as excellent candidates for the IMTA land-based systems.
ACKNOWLEDGE
These results were obtained within the framework of the ERA-Net COFASP project (IMTA-Effect project) with funding from Fundação para a Ciência e Tecnologia (COFASP/0003/2015).
REFERENCES
- Kraft LG, Kraft GT, Waller RF (2010) Investigations into Southern Australian Ulva (Ulvophyceae, Chlorophyta) taxonomy and molecular phylogeny indicate both cosmopolitanism and endemic cryptic species†. Journal of phycology 46: 1257-1277.
- Ben-Ari T, Neori A, Ben-Ezra D, Shauli L, Odintsov V, et al. (2014) Management of Ulva lactuca as a biofilter of mariculture effluents in IMTA system. Aquaculture 434: 493-498.
- Carl C, De Nys R, Paul NA (2014) The seeding and cultivation of a tropical species of filamentous Ulva for algal biomass production. PLoS One 9: 98700.
- Ding L, Teng L, Lu Q, Luan R, Huang B (2014) The morphological comparison, variation and molecular analysis between two green tidal algae Enteromorpha prolifera and clathrata from China. Biology.
- Bruhn A, Dahl J, Nielsen HB, Nikolaisen L, Rasmussen MB, et al. (2011) Bioenergy potential of Ulva lactuca: Biomass yield, methane production and combustion. Bioresource Technology 102: 2595-2604.
- Castelar B, Reis RP, dos Santos Calheiros AC (2014) Ulva lactuca and flexuosa (Chlorophyta, Ulvophyceae) cultivation in Brazilian tropical waters: Recruitment, growth, and Ulvan yield. Journal of Applied Phycology 26: 1989-1999.
- Costea O, Malta E-J, Lópeza JC, Fernández-Díaza C (2015) Production of sulfated oligosaccharides from the seaweed Ulva using a new Ulvan-degrading enzymatic bacterial crude extract. Algal Research 10: 224-231.
- Winberg PC, Skropeta D, Ullrich A (2011) Seaweed cultivation pilot trials-towards culture systems and marketable products. Australian Government Rural Industries Research and Development Corporation, Canberra, Australia.
- Radulovich R, Neori A, Valderrama D, Reddy CRK, Cronin H, et al. (2015) Farming of seaweeds. Seaweed Sustainability: Food and Non-Food Applications. Pg no: 27-59.
- Melton JT, Collado-Vides L, Lopez-Bautista JM (2016) Molecular identification and nutrient analysis of the green tide species Ulvaohnoi Hiraoka& S. Shimada, 2004 (Ulvophyceae, chlorophyta), a new report and likely nonnative species in the Gulf of Mexico and Atlantic Florida, USA. Aquatic Invasions 11: 225-237.
- Shimada S, Hiraoka M, Nabata S, Iima M, Masuda M (2003) Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycological Research, 51: 99-108.
- Hofmann L, Nettleton J, Neefus C, Mathieson AC (2010) Cryptic diversity of Ulva (Ulvales, Chlorophyta) in the Great Bay Estuarine System (Atlantic USA): Introduced and indigenous distromatic species. European Journal of Phycology 45: 230-239.
- Cormaci M, Furnari G, Alongi G (2014) Flora marina bentonica del Mediterraneo: Chlorophyta. Bollettinodell’Accademia Gioenia Di Scienze Naturali 47: 11-436.
- Loughnane CJC, McIvor LLM, Rindi F, Stengel DB, Guiry MD (2008) Morphology, rbcL phylogeny and distribution of distromatic Ulva (Ulvophyceae, Chlorophyta) in Ireland and southern Britain. Phycologia 47: 416-429.
- Heesch S, Broom JES, Neill KF, Farr TJ, Dalen JL, et al. (2009) Ulva, UmbraUlva and gemina: Genetic survey of New Zealand taxa reveals diversity and introduced species. European Journal of Phycology 44: 143-154.
- Mareš J, Leskinen E, Sitkowska M, Skácelová O, Blomster J (2011) True identity of the european freshwater Ulva (Chlorophyta, Ulvophyceae) revealed by a combined molecular and morphological approach. Journal of Phycology 47: 1177-1192.
- Wolf MA, Sciuto K, Andreoli C, Moro I (2012) Ulva (Chlorophyta, Ulvales) Biodiversity in the north adriatic sea (Mediterranean, Italy): Cryptic species and new introductions. Journal of Phycology 48: 1510-1521.
- Lawton RJ, Mata L, de Nys R, Paul NA (2013) Algal Bioremediation of waste waters from land-based aquaculture using Ulva: Selecting target species and strains. PLoS One 8: 77344.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc Biol Sci 270: 313-321.
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102: 8369-8374.
- Lin Z, Shen S, Chen W, Li H (2013) Phylogenetic analyses of four species of Ulva and Monostroma grevillei using ITS, rbc L and 18S rDNA sequence data. Chinese Journal of Oceanology and Limnology 31: 97-105.
- Rybak A, Czerwoniec A, G?bka M, Messyasz B (2014) Ulvaflexuosa (Ulvaceae, Chlorophyta) inhabiting inland aquatic ecosystems: Molecular, morphological and ecological discrimination of subspecies. European Journal of Phycology 49: 471-485.
- Shimada S, Yokoyama N, Arai S, Hiraoka M (2008) Phylogeography of the genus Ulva(Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. In: Borowitzka MA, Critchley AT, Kraan S, Peters A, Sjøtun K, Notoya M (eds.). Nineteenth International Seaweed Symposium. Developments in Applied Phycology, Springer, Dordrecht, Netherlands.
- Saunders GW, Kucera H (2010) An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie Algologie, 31: 487-528.
- Gao G, Zhong Z, Zhou X, Xu J (2016) Changes in morphological plasticity of Ulva prolifera under different environmental conditions: A laboratory experiment. Harmful Algae 59: 51-58.
- Vila-Concejo A, Ferreira Ó, Matias A, Dias JMA (2003) The first two years of an inlet: Sedimentary dynamics. Continental Shelf Research 23: 1425-1445.
- White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. PCR - Protocols and Applications - A Laboratory Manual, Publisher: Academic Press, Massachusetts, USA. Pg no: 315-322.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647-1649.
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ (2017) Nucleic Acids Research 45: 37-42.
- Madden T (2002) The blast sequence analysis tool. The NCBI Handbook, Maryland, USA.
- Katoh K, Toh H (2008) Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics, 9: 212.
- Mareš J (2011) Combined morphological and molecular approach to the assessment of Ulva (Chlorophyta, Ulvophyceae) in the CzechRepublic. University of South Bohemia, Faculty of Science, Department of Botany, Bohemia, Czech Republic. Pg no: 76.
- Guindon S, Gascuel O, Rannala B (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696-704.
- Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: More models, new heuristics and parallel computing. Nat Methods 9: 772-772.
- Phillips JA, Lawton RJ, Denys R, Paul NA, Carl C (2016) Ulvasaporasp., an abundant tubular species of Ulva (Ulvales) from the tropical pacific ocean. Phycologia, 55: 55-64.
- Masakiyo Y, Shimada S (2014) Species diversity of the genus Ulva (Ulvophyceae, Chlorophyta) in Japanese waters, with special reference to Ulvatepida Masakiyo et S. Shimada nov. Bull Natl Mus Nat Sci Ser B 40: 1-13.
- Couceiro L, Cremades J, Barreiro R (2011) Evidence for multiple introductions of the Pacific green alga Ulva australis Areschoug (Ulvales, Chlorophyta) to the Iberian Peninsula. Botanica Marina 54: 391-402.
- O’Kelly CJ, Kurihara A, Shipley TC, Sherwood AR (2010) Molecular assessment of Ulva (Ulvophyceae, Chlorophyta) in the Hawaiian islands. Journal of Phycology 46: 728-735.
- Wynne MJ (2005) A check-list of benthic marine algae of the tropical and subtropical Western Atlantic: Second revision. Beiheftezur Nova Hedwigia, California, USA. Pg no: 152.
- Sfriso A, Curiel D (2007) Check-list of seaweeds recorded in the last 20 years in Venice lagoon, and a comparison with the previous records. Botanica Marina, Walter de Gruyter, Berlin, Germany. Pg no: 22-58.
- Guiry MD, Guiry GM (2018) World-wide electronic publication. Algaebase, Galway, Ireland.
- Rybak AS (2018) The Ulva flexuosa complex (Ulvaceae, Chlorophyta): An updated identification key with special reference to the freshwater and hyperhaline taxa. Phytotaxa 345: 83-103.
- Neori A, Chopin T, Troell M, Buschmann AH, Kraemer GP, et al. (2004) Integrated aquaculture: Rationale, evolution and state of the art emphasizing seaweed biofiltration in modern mariculture. Aquaculture 231: 361-391.
- Lee SH, Kang PJ, Nam KW (2014) New record of two marine ulvalean species (Chlorophyta) in Korea. Journal of Ecology and Environment37: 379-385.
- Hayden HS, Blomster J, Maggs CA, Silva PC, Stanhope MJ, et al. (2003) Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. European Journal of Phycology 38: 277-294.
- Wichard T, Charrier B, Mineur F, Bothwell JH, De Clerck O, et al. (2015) The green seaweed Ulva: A model system to study morphogenesis. Front Plant Sci 7: 6-72.
- Grueneberg J, Engelen AH, Costa R, Wichard T (2016) Macroalgal morphogenesis induced by waterborne compounds and bacteria in coastal seawater. PLoS One 11: 0146307.
- Jones AB, Dennison WC, Preston NP (2001) Integrated treatment of shrimp effluent by sedimentation, oyster filtration and macroalgal absorption: A laboratory scale study. Aquaculture, 193: 155-178.
- Benhissoune S, Boudouresque CF, Verlaque M (2001) A check-list of marine seaweeds of the Mediterranean and Atlantic coasts of Morocco. Chlorophyceaewille s. l. Botanica Marina 44: 171-182.
- Hernández I, Bermejo R, Lucas J, Juan P, Vergara J, et al. (2010) Contribucion al conocimiento de los macrófitos marinos de saco interno y caños adyacentes de la bahia de Cadiz. Algas 43: 11-16.
- Araújo R, Bárbara I, Tibaldo M, Berecibar E, Tapia PD, et al. (2009) Checklist of benthic marine algae and cyanobacteria of northern Portugal. Botanica Marina 52: 24-46.
- Peña V, Barbara I (2002) Caracterización florística y zonación de las algas bentónicas marinas del Puerto de a coruña (N.O. Península Ibérica). Nova Acta Cientifica Compostelana (Bioloxía)12: 35-66.
- Aníbal J, Madeira HT, Carvalho LF, Esteves E, Veiga-Pires C, et al. (2014) Macroalgae mitigation potential for fish aquaculture effluents: An approach coupling nitrogen uptake and metabolic pathways using Ulvarigida and Enteromorpha clathrata. Environmental Science and Pollution Research 21: 13324-13334.
- Alsufyani T, Engelen AH, Diekmann OE, Kuegler S, Wichard T (2014) Prevalence and mechanism of polyunsaturated aldehydes production in the green tide forming macroalgal genus Ulva (Ulvales, Chlorophyta). Chemistry and Physics of Lipids 183: 100-109.
- Fleming T (2016) Analysis of freshwater and marine Ulva flexuosa in Southern California. Pg No: 1-28.
- Zhao J, Jiang P, Qin S, Liu X, Liu Z, et al. (2015) Genetic analyses of floating Ulva prolifera in the Yellow Sea suggest a unique ecotype. Estuarine, Coastal and Shelf Science 163: 96-102.
- Zhang Q (2015) Porphyra aquaculture rafts is the major source of floating green algae in the yellow sea: Evidence of intraspecific genetic analysis on Ulva prolifera. National Center for Biotechnology Information, Maryland, USA.
- Ogawa T, Ohki K, Kamiya M (2013) Differences of spatial distribution and seasonal succession among Ulva species (Ulvophyceae) across salinity gradients. Phycologia 52: 637-651.
Citation: Favot G, Engelen AE, Cunha ME, Serrão MEA (2019) Identification of Ulva sp. Grown in Multitrophic Aquaculture Systems. J Aquac Fisheries 3: 024.
Copyright: © 2019 Glauco Favot, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

