Introduction
Rice (Oryza sativa L.) is the most important staple crop that feeds nearly half of the world’s population. Because originated in tropical or subtropical areas, rice is sensitive to low temperatures throughout its entire life cycle. Cold stress can inhibit photosynthesis by reducing the chlorophyll concentration, inducing the accumulation of Reactive Oxygen Species (ROS) and by causing severe damage to various cellular components including membrane lipids, structural proteins and enzymes [1-3]. Low temperatures have the potential to negatively affect the growth and development of rice plants during any developmental stage, from germination to grain filling, resulting in large losses in rice production [4].
Plants have developed several strategies to respond to low temperatures that are known collectively as ‘cold acclimation’ [5]. Numerous studies of cold acclimation have been reported in recent years, and they showed that many pathways participate in the cold-response process [6,7]. The Dehydration-Responsive Element Binding protein (DREB1)/C-repeat binding factor (CBF)-dependent cold-responsive pathway is the most well-known among all the pathways [8]. Another pathway with a central role in the rice response to cold stress is the MYBS3-dependent pathway. Interestingly, DREB1A responds early and transiently, while MYBS3 responds relatively slowly to cold stress in rice, suggesting that theMYBS3-dependent pathway is important for long-term adaptation to persistent cold stress [9]. In addition to these two pathways, the plant hormone systems play an indispensable role in cold stress tolerance [10]. For example, Jasmonate (JA) positively regulates cold tolerance in plants because it can positively modulate the CBF-dependent or –independent pathway, leading to accumulation of cryoprotective compounds such as polyamine, glutathione and anthocyanins [11]. DELLA proteins, the key components of the Gibberellic Acid (GA)-signaling pathway, are involved in the CBF1-mediated cold stress response [12]. The cross-talk between Cytokinin (CK) and Abscisic Acid (ABA) are also important in cold stress signaling. Components of CK signaling, Histidine Kinases (AHKs) and the cold-inducible A-type ARRs, play negative regulatory roles in cold stress response via inhibition of the ABA response [13]. Components of Brassinosteroid (BR) and ethylene signaling were also found to modulate freezing tolerance in Arabidopsis plants [14,15].
There are two major subspecies of rice, Oryza sativa ssp. Indica and Oryza sativa ssp. japonica. Generally, japonica rice varieties are more tolerant to low temperatures than varieties of indica rice [16]. Therefore, cold tolerance in indica rice can be improved by introducing genes from japonica rice through molecular breeding. Recent advances in next generation sequencing technology provide a powerful strategy for discovering new genes associated with cold tolerance in rice [17-21]. In this study, we combined transcriptome sequencing with genome re-sequencing to identify the Cold-Responsive-genes (CORs) that are differentially-expressed between the two rice subspecies. Using this approach, we found four COR genes that are differentially-expressed between indica and japonica varieties. Moreover, these genes show important natural variation between the two subspecies which predicted to result in changes of cold responsive cis-elements. Therefore, these CORs may be useful for improving cold tolerance in rice through molecular breeding.
Results
Global mRNA expression profiles in rice seedlings in response to cold stress
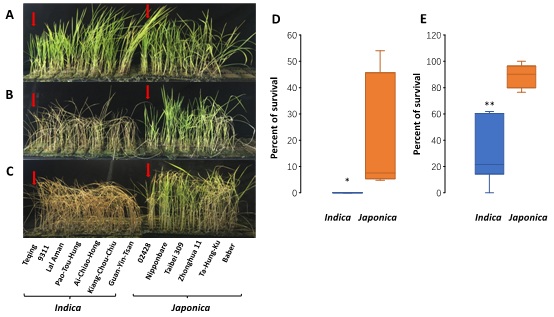
In general,
japonica rice varieties display a higher potential for cold tolerance than do
indica varieties [16] (Figure S1). In order to fully understand the genetic basis of the difference in the cold response between the two subspecies, we subjected seedlings of two cultivars, Teqing and 02428, to low temperature treatments at 4°C and 10°C. As shown in figure S1, the seedlings showed contrasting cold tolerance in response to the two treatments, suggesting that these two cultivars, Teqing and 02428 are representative of the cold sensitivity of
indica rice and the cold tolerance of
Japonica rice.
A total of 10 sequencing libraries were constructed from RNA extracted from the shoot tissues of three-leaf stage rice seedlings of the
indica rice cultivar Teqing and the
japonica cultivar 02428exposed to low temperature conditions(4°C and 10°C) for 0hr, 3hr and 24hr. Because there was no difference between the two 0-hr cold treatment samples, two common controls, I0 and J0, were used for the low temperature treatments (4°C and 10°C) in the
indica and
japonica rice cultivars, respectively.
The Solexa Genome Analyzer was used to perform high-throughput Tag-seq analysis on these libraries. As shown in table 1, we obtained5.8 to 6.2 million raw sequence tags per library, of which 251, 288 to 301,590 tag sequences were distinct. After filtering out the 3' adaptor sequence, empty reads, low-quality tags, unexpected-length tags and single-copy tags, approximately 5.7 to 6.0 million clean tags remained in each sample. Finally, we obtained 111,174 to 136,023 unique tags for each of the ten libraries. Saturation analysis was applied to estimate whether or not new unique tags can be detected with increases in the total number of tags. The numbers of unique tags increased with the total number of tags and reached a plateau shortly after 1 million tags; no new unique tag was identified as the total number of tags approached 2 million. Therefore, the ten libraries are full representations of transcripts under the different treatments.
We then annotated the sequence tags based on the Os-Nipponbare-Reference-IRGSP-1.0 genome assembly (
http://rice.plantbiology.msu.edu) and only clean and unambiguous tags that matched perfectly or had only a single mismatch were analyzed further [22]. Based on these criteria, 45.50%-49.31% of the clean tags in all the ten libraries mapped to unique positions on the reference genes. 8.27%-12.59% of the clean tags from these same libraries were matched to the reference genome database. However, 3.67%-8.54% of the clean tags database from the 10 libraries did not map to the reference genome.
Identification of COR genes in the indica and japonica rice cultivars
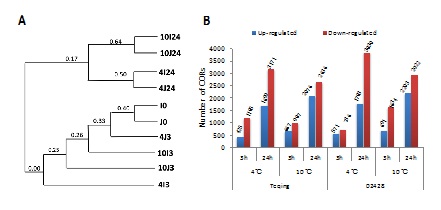
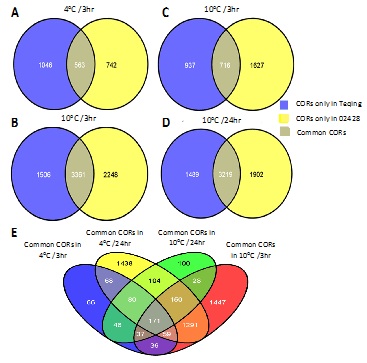
Based on hierarchical clustering of the 10 transcriptomes, the 10 experimental samples were classified into two clades (Figure 1). The first clade contained the samples exposed to the low temperature treatments (4 or 10°C) for a short time (3hr) and the 0-hr controls without cold treatment, while the second clade contained the samples exposed to the cold treatments (4 or 10°C) that lasted for 24hr. Moreover, in the second clade, there are two sub-clades that correspond to the two temperatures, and each sub-clade contained both indica and japonica rice cultivars. These results suggest that the differences between the duration of cold treatment were more significant than the differences between either the two low temperature treatments or between the two cultivars, and that the cultivar difference was the smallest.
In order to find CORs in the two cultivars, we compared gene expression between cold treatment samples and the control samples for the same cultivar. The transcripts with at least two-fold differences (|log2Ratio| ≥1 and FDR ≤ 0.001) were referred as CORs. As shown in figure 1, there were a total of 1610 CORs in Teqing and 1307 CORs in 02428 under the 4°C/3hr treatment. After 3-hour exposure to 10°C, 1654 CORs were expressed in Teqing, and 2325 CORs were expressed in 02428. Following 24-hour cold treatment, more CORs were identified in these two cultivars: for the 4°C/24hr treatment, 4870 and 5613 CORs were expressed in Teqing and 02428, respectively, while there were 4712 and 5125 CORs expressed in Teqing and 02428 under the 10°C/24hr treatment, respectively.
Gene ontology and pathway enrichment analysis of the rice CORs
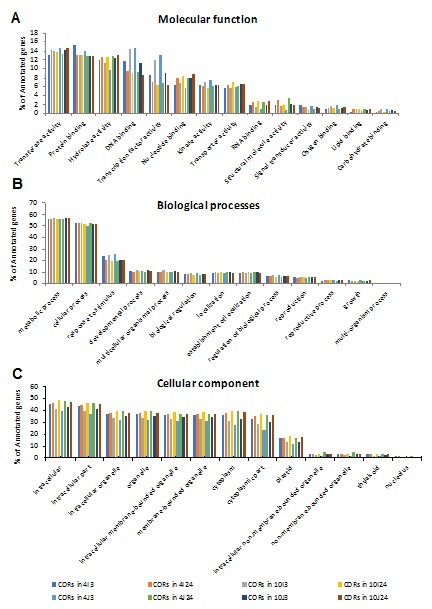
In order to reveal functions of the CORs identified above, we performed Gene Ontology (GO) enrichment analysis. As shown in figure 2, the CORs in the two cultivars were enriched in the “molecular function” terms “transferase activity”, “protein binding”, “hydrolase activity”, “DNA binding”, “protein binding” and “hydrolase activity”. The CORs in Teqing and 02428 were both enriched in the “biological process” terms “metabolic process” and “cellular process”. Genes involved in “intracellular organelle”, “membranes” and “cytoplasm” were also significantly enriched among the “cellular component” GO terms. However, there were no GO terms enriched only in Teqing or 02428.
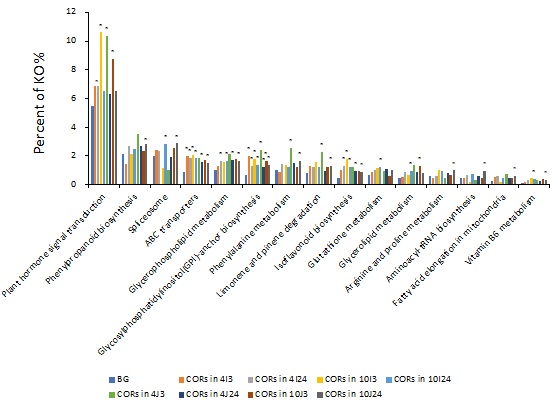
According to the KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis, there were 15 significantly enriched KEGG pathways in both cultivars (Figure 3), included “plant hormone signal transduction”, “phenylpropanoid biosynthesis”, “spliceosome”, “ABC transporters”, “glycerophospholipid metabolism”, “Glycosylphosphatidylinositol (GPI)-anchor biosynthesis”, “phenylalanine metabolism”, “limonene and pinene degradation”, “isoflavonoid biosynthesis”, “glutathione metabolism”, “glycerolipid metabolism”, “arginine and proline metabolism”, “aminoacyl-tRNA biosynthesis”, “fatty acid elongation in mitochondria” and “vitamin B6 metabolism”. Among these pathways, the pathway with the most significant representation was “plant hormone signal transduction”. These results suggest that plant hormones could play an important role in regulating cold tolerance in rice.
Identification of consistent differentially-expressed COR genes in multiple rice cultivars
CORs that are present in many rice varieties and that respond to different low temperature conditions may be key genes in cold response pathways. To screen such genes, we used Venn diagrams to classify CORs in Teqing and 02428. Based on the results shown in figure 4, we conducted a further Venn diagram analysis and found a total of 171 CORs that were shared by both cultivars and consistently responded to the four different cold treatments. Thus, they may represent crucial cold responsive genes that act in specific pathways.
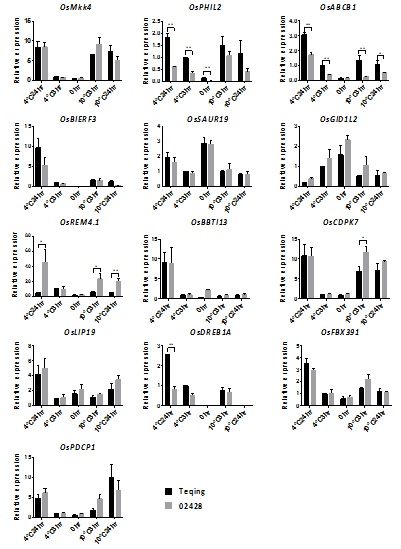
To find the key genes representing the specific pathways, we screened out 13 of the 171 genes we found that were included in predicted cold-response pathways (
Supplemental Data 1). To confirm the cold-induced expression of these genes, gene-specific primers were designed for all13 genes and qRT-PCR assays were performed (Figure 5). The expression patterns of these 13 genes agreed with the RNA-seq results, confirming the high quality of the datasets. Among these genes,OsABCB5(LOC_Os01g50100, encoding a member of ABCB subgroup 5 in ABC transporter super family), OsPHIL2 (LOC_Os02g52010, encoding a member of Phi-1-like phosphate induced protein) and OsDREB1A (LOC_Os09g35030, encoding a member of the CBF/DREB1 protein family) had higher expression levels in Teqing than in 02428, while OsCDPK7 (LOC_Os04g49510, encoding a member of calcium-dependent protein kinase family) and OsREM4.1 (LOC_Os07g38170, encoding a member of the remorin family) had significantly lower expression levels in Teqing than in 02428 in response to the cold treatments.
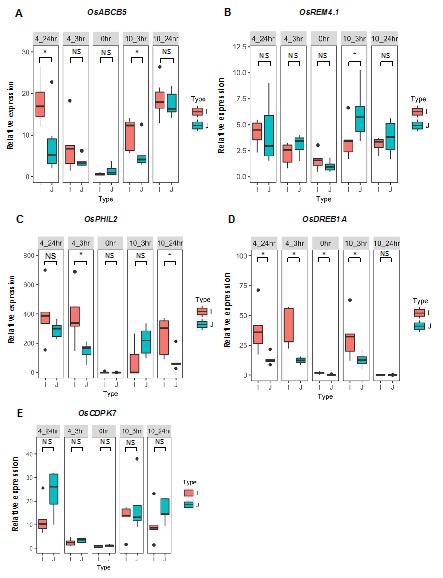
To determine whether the observed differences in expression for the five genes are associated with cold tolerance in other cultivars of the two subspecies, we included an additional five cold tolerant
japonica rice cultivars (Nipponbare, Ta Hung Ku, Taibei309, Baber, Zhonghua 11) and five cold sensitive
indica rice cultivars (9311, Lal-Aman, Pao-Tou-Hung, Ai-Chiao-Hong, Guan-Yin-Tsan) to the study. Details of the cold responses of these 10cultivars are shown in figure S1. All of the
japonica rice cultivars showed significantly stronger cold tolerance than the
indica cultivars, as expected. We then we assayed the expression levels of the five genes (OsABCB5, OsPHIL2, OsDREB1A, OsCDPK7 and OsREM4.1) in the 10 cultivars. Among these five genes, the expression profiles of OsABCB5, OsPHIL2, OsDREB1A and OsREM4. 1still showed clear distinctions between the
indica and
japonica subspecies, while OsCDPK7 did not show the
indica-
japonica difference in the multi-cultivar comparison (Figure 6). In the initial comparison between Teqing and 02428, OsABCB5, OsPHIL2 and OsDREB1Awere expressed at higher levels in Teqing than in 02428; similarly, in the comparison of the five cultivar pairs, the expression of the three genes was also higher in
indica than in
japonica. OsREM4.1 showed higher expression levels in the
japonica rice cultivars, confirming the results from the comparison of Teqing and 02428. After cold treatment, expression of OsABCB5, OsPHIL2, OsDREB1A and OsREM4. 1 was significantly higher than in the control, and the expression of OsPHIL2 was up-regulated >400-fold after 4°C treatment in the
indica cultivars.
Analysis of natural variation in these COR genes in multiple rice cultivars
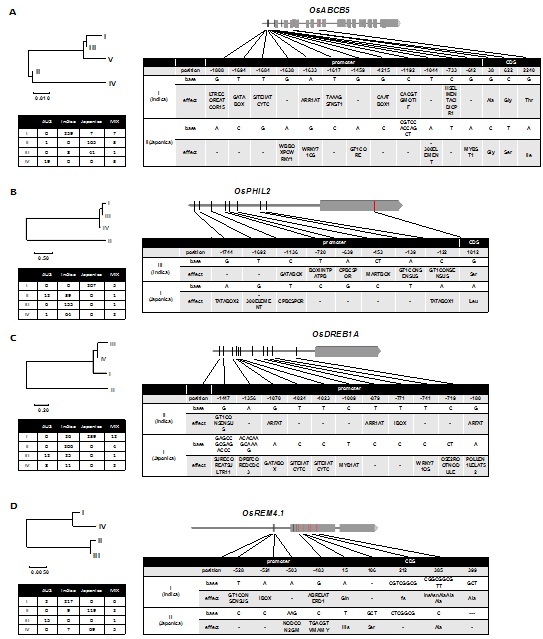
To investigate the natural variations in the sequences of OsABCB5, OsPHIL2, OsDREB1A and OsREM4.1, we extracted the sequences of their 2.0-kb promoter regions and coding regions from the re-sequencing dataset of a diversity panel of 575 rice cultivars that included 294 indica and 239 japonica lines. The original sequencing datasets have been deposited in the Genome Sequence Archive of the Bejing Institute of Genomics, Chinese Academy of Sciences (bigd.big.ac.cn/gsa) under Accession numbers CRA000778, CRA000779 and CRA000995 (Figure 7).
Based on nucleotide polymorphisms, we could divide the sequences of the 575 lines into four haplotypes for OsABCB5; among these, HapII was the most represented in japonica lines, and Hap I was the most represented in indica lines. There were 12 SNP/Indel variations identified in the OsABCB5 promoter region between indica and japonica lines. Of these, four were predicted to be located within low temperature responsive cis-elements. For example, Hap II and Hap I differed in the nucleotides located at positions -642 G/A, -1633 A/G, -1638 G/A, and -1808 G/A upstream of the start codon; these four SNPs were related to theMYBST1, WRKY71OS/ARR1AT, WBBOXPCWRKY1 and LTRECOREATCOR15motifs, respectively, that are predicted to function in the cold response.
Based on the 79 polymorphic nucleotide sites, a total of five haplotypes of OsPHIL2were identified; among them, Hap I and Hap III were unique to japonica and indica rice lines, respectively. In the promoter region of OsPHIL2, we found 8SNP variations between Hap I and Hap III, and of these, no SNP was related to the cold response directly, but both -639A/G and -1136C/T were predicted to be associated with cytokinin.
By analyzing the candidate regulatory region of OsDREB1A in the 575 accessions, we found four haplotypes; Hap I and Hap II were unique to japonica and indica lines, respectively. In the promoter region, we identified 11SNPs/In Dels, among them, seven SNPs are located at -180, -741, -771, -720, -879, -1009 and -1070bp upstream of the TSS (transcription start site), and correspond to G-A, T-C, T-C, T-C, T-C, T-C, C-T and G-A changes, respectively. The single In Delis 11bpin length (CACAAGCAAAG) and is located 1346-1356bp upstream of the TSS. The SNPs located at positions -180 and -1070 were predicted to be related to auxin, SNPs located at -771 and -1009 were predicted to be related to MYB, SNPs at -741, -879, and the In Del were related to WRKY71OS, ARR1AT,and MYB1AT, respectively. Among these motifs, WRKY71OS and MYB1AT were related to MYB while ARR1AT was related to cytokine in.
We analyzed the promoter and CDS region of OsREM4.1by DNA sequencing and identified 48 variations. Analysis of these variations indicated that two major haplotypes were present in the 575 accessions; Hap I was present in 217 indica rice lines, and Hap II was present in 119 japonica rice lines. We found four In Dels and five SNPs in the candidate regulatory region. There is a SNP located at -483upstream of the TSS, corresponding to a G-C change and related to the ABRELATERD1motifwhichis related to ABA.
Discussion
The japonica subspecies of Oryza sativa has adapted to a temperate climate, while the indica subspecies has adapted to tropical and subtropical environments. As a result, japonica rice cultivars are generally more tolerant to cold stress than indica rice cultivars (Figure S1). In our study, we used comparative transcriptome analysis on two rice cultivars to identify a group of COR genes in these two cultivars. From the CORs, we further identified four key genes, OsABCB5, OsPHIL2, OsDREB1A and OsREM4.1, that showed ubiquitous and distinctive activity in indica and japonica cultivars in an integrative study using KEGG and GO annotations, Venn diagram analysis, qRT-PCR, and natural variation analysis.
The DREB1/CBF pathway is a well-known cold-responsive pathway in plants [23]. The pathway includes the CBF1, CBF2 and CBF3 genes, which are also known as DREB1B, DREB1C and DREB1A, respectively, in Arabidopsis [24]. However, not all three transcription factors act as positive regulators in the CBF/DREB1 pathway. In Arabidopsis, freezing tolerance of the cbf2 mutant is enhanced by up-regulating the expression of CBF1/DREB1B and CBF3/DREB1A, which means that CBF2/DREB1C plays a negative role in the cold response [25]. OsDREB1A has been reported previously to be induced by cold stress [26,27]. Consistent with these results, we found that the expression of OsDREB1Aincreased after cold treatment. Lourenco et al., have claimed that in OsHOS1 knockdown plants, the reduced expression of OsHOS1 promoted the accumulation of OsICE1 protein and resulted in the up-regulation of OsDREB1A. However, the transgenic plants did not show increased cold tolerance [28]. In our study, the expression level of OsDREB1A was higher in indica rice compared to japonica rice. Therefore, we speculate that OsDREB1Amay play a negative role in cold signaling. In our natural variation analysis, we discovered a SNP related to MYB in the promoter region of OsDREB1Ain HapI (japonica). Interestingly, a MYB-like transcription factor has been reported to be involved in the cold regulation of CBF genes, and MYB15 over expression reduced the expression of CBF genes under cold treatment [29]. Thus, the MYB negative regulation of OsDREB1A may result in the stronger cold tolerance observed in japonica rice cultivars.
Among the four genes we screened, OsREM4.1 is related to the plant hormone ABA, which is known as a cold stress-responsive hormone. Two SNP variations within ABA responsive or MYB elements were located within the promoter region of OsREM4.1. Interestingly, OsREM4.1 was previously characterized and found to be up-regulated by ABA signaling; the OsREM4.1 protein can bind to OsSERK1 and inhibit the formation and activation of the OsBRI1-OsSERK1 receptor complex, which is crucial for BR signaling [30]. In Arabidopsis thaliana, application of exogenous ABA can enhance cold tolerance and induce the expression of several COR genes, including RAB18, RD29A and KIN2 [31,32]. Several studies showed that plants treated with BR grew better at low temperature compared to optimal conditions [33]. In addition, Li et al., described two key components of BR signaling, BIN2 and BZR1, regulate plant freezing tolerance; BIN2 and its homologs play negative roles, while BZR1 plays a positive role in the cold response [14]. In our study, OsREM4.1hada higher level of expression in japonica rice, so we predicted that OsREM4.1 may play a positive role in regulating cold tolerance via crosstalk between the ABA and BR pathways.
Another gene we found that is associated with the BR pathway is OsPHIL2, which encodes a Phosphate-Induced protein (PHI). Interestingly, two BR-response genes in Arabidopsis, EXORDIUM (EXO) and EXORDIUM-LIKE1 (EXL1), were previously reported to have the same PHI conserved region [34]. Expression of the EXO gene is up-regulated by BR and EXL1 has a similar expression pattern to EXO in Arabidopsis in that it can be induced by BR [35,36]. We therefore predicted that OsPHIL2 may be similarly connected to BR signaling pathway. Considering that OsPHIL2 is expressed at higher levels in indica cultivars when exposed to low temperature, OsPHIL2 may negatively regulate cold stress via BR signaling.
ABC transporters constitute one of the largest protein families, and are present in organisms ranging from bacteria to humans [37]. In plants, ABC proteins were originally identified as transporters involved in the final detoxification process [38]. OsABCB5 is predicted to encode an MDR-like ABC transporter. There are several studies that have reported that MDR-like ABC transporters are involved in the auxin signaling pathway. Noh et al., showed that two MDR-like genes, At MDR1 and AtPGP1, are required for auxin transport and auxin-mediated development in Arabidopsis. Also, mutation of two related MDR-like genes (MDR1 and PGP1) resulted in reduced polar auxin transport [39]. Another study found that the Arabidopsis MDR-like ABC transporter4 (AtPGP4) is involved in auxin-mediated root development [40]. Although the role of auxin in cold stress remains unclear, there are reports that potentially link cold stress to auxin by showing that cold stress inhibits the inflorescence gravity response in Arabidopsis thaliana [41,42]. An early study demonstrated that temperature affects the speed of exogenous auxin transport in a variety of plant species [43]. Considering that auxin is at the center of hormonal crosstalk, auxin may mediate the response to cold by interacting with other hormones [44]. These results indicate that OsABCB5may play a crucial role in the cold response in rice seedlings by transporting auxin.
Based on the expression profiles and predicted pathways of the four key genes involved in the cold response in rice, we can conduct further functional studies to determine the biological and biochemical functions of the proteins encoded by these genes.
Materials and Method
Plant materials and Evaluation of Phenotypes
In this study, we used Teqing and 02428 as the cold-sensitive indica and cold-tolerant japonica cultivars, respectively. We also chose an additional 5 japonica and 6 indica cultivars for cold temperature treatment. Seeds were allowed to germinate for 3 days in distilled water, after which they were transferred to moist filter paper. The seedlings were grown in 0.5-fold Kimura B solution at pH 6.0 under a 13-h light (28°C)/11-h dark (25°C) photoperiod and 70-80% relative humidity [45]. Three-leaf rice seedlings were transferred to a climatic chamber (Percival Scientific, Perry, IA) and maintained at either 4°C or 10°C or for 4 days and 5 days, respectively. After the plants had recovered at 28°C /25°C day/night cycles for 7 days, the survival rates were determined based on the percentage of surviving seedlings [46]. All treatments were repeated at least three times, and the average of the replicates was used to characterize cold tolerance. The significant differences between subspecies were analyzed using ANOVA or Student’s t-test.
RNA isolation
Total RNA was isolated from plant tissue using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA). The RNA quality was assessed visually by electrophoresis on agarose gels, and the concentration was determined with a NanoDrop spectrophotometer (ND-1000, NanodropTechnologies, Wilmington, DE, USA).
Library construction and Illumina cDNA sequencing
Whole transcriptome analysis was performed using RNA isolated from three-leaf rice seedlings that were cold-treated at 10°C or 4°C for 0, 3 and 24 hours under continuous illumination. We extracted total RNA, used oligo (dT) magnetic beads to purify the mRNA, and then used Oligo (dT) as a primer to synthesize cDNA. The 5' ends of the cDNA tags can be generated with two different restriction end nucleases, NlaIII or DpnII, Usually, the bead-bound cDNA fragments are digested with NlaIII, which recognizes and cuts atthe CATG sites. The 3' cDNA fragments connected to the oligo (dT) beads were washed away and the Illumina adaptor 1 was ligated to the 5'sticky ends of the digested bead-bound cDNA fragments. The junction of the Illumina adaptor 1 and the CATG site is an MmeI recognition site, and it cuts 17bp downstream of the CATG site, producing tags containing adaptor 1. After removing the 3' fragments with magnetic beads precipitation, Illumina adaptor 2 was ligated to the 3' ends of the tags, resulting in tags with different adaptors on each end to form a tag library. Following linear PCR amplification of the sample libraries, the double-stranded cDNA fragments are purified by polyacrylamide gel electrophoresis prior to high-throughput cDNA sequencing using the Illumina HiSeqTM 2000 instrument.
Identification of COR genes
The genes with differential expression between 0 hours and 3/24 hours of cold treatments are Called Cold Responsive genes (CORs). Before comparing the differential expression of genes in response to cold treatment, normalized gene expression levels were obtained by normalizing the number of raw clean tags in each library to the number of Transcripts Per Million clean tags (TPM). A rigorous algorithm method was performed to detect differential expression of genes across samples. A combination of FDR ≤ 0.001 and the absolute value of log 2 Ratio ≥1 were used as the threshold to determine the significance of differentially-expressed genes. GO and pathway enrichment analyses were based on the agriGO (
http://bioinfo.cau.edu.cn/agriGO/index.php) and KEGG pathway databases (
http://www.genome.jp/kegg/) [47-49]. Cluster analysis was performed with CLUSTER3.0 and viewed using the TREEVIEW software program (
http://rana.lbl.gov/EisenSoftware.htm).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of gene expression
An independent experiment was performed to examine the expression of putative COR genes using qRT-PCR. Total RNA was extracted from rice leaves before and after exposure to 4°C or 10°C for 3h and 24h using the TRIzol Reagent (Invitrogen).cDNA was synthesized and qRT-PCR was performed according to the previous report [50]. All reactions were performed with three biological and three technical replicates. OsActin was used as an internal control. The sequences of primers used for the qRT-PCR assays are listed in table S1. The gene expression levels were calculated using the 2-∆∆Ct method. Student’s t-test was used to determine whether there were significant differences between samples.
Haplotype analysis and SNP discovery of the four key COR genes
Haplotype analysis was performed on the genomic sequences that included the OsABCB5, OsPHIL2, OsDREB1A and OsREM4. 1 promoter and coding regions from the 575 rice accessions. A neighbor-joining tree based on the nucleotide sequences of the four key genes from the rice varieties was constructed using MEGA 5.0.
For a genome-wide association study, we used our own SNP database to performed SNP analysis. Additionally, we used the Genome Information Database System (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi) to predict the functions of the SNPs that we found.
Author Contribution
DM conceived and designed the experiments. XH and DM performed the experiments. XH and YT analyzed the data, and XH wrote the manuscript.
Acknowledgement
We thank Dr. Caiyan Chen for his help in conceiving the project. This work was supported by grants from the National Natural Science Foundation of China (31101211, 31371603).