ABSTRACT
MicroRNAs (miRNAs) are small non-coding single-stranded RNA molecules (containing about 22 nucleotides) that are widely found in many organisms, as varied as micro-organisms, plants, invertebrates and vertebrates. These miRNAs function in RNA silencing and post-transcriptional regulation of gene expression. MiRNAs have been found to be involved in a number of biological and pathological processes, including sperm biology and infertility. This review will describe and discuss the role of miRNAs in terms of their role in sperm function, male infertility and gene regulation.
KEYWORDS
Infertility; MicroRNAs; Non-coding RNA; Sperm; Transgenerational inheritance
Introduction
The sperm cell has become highly specialised, with the sole purpose to fertilize an oocyte. Sperm have a single purpose, which is to fertilize the oocyte - and with this it has become a highly complex and specialised cell. The structure of the sperm assumes a simplistic structure, consisting of a head, which is mostly void of cytoplasm, a nucleus packed with DNA tightly wound around protamines; a midpiece housing the mitochondria necessary to fuel its locomotion, and a tail or principle piece that whips in motion to move the sperm towards the oocyte. However, despite the lack of cytoplasmic space, increasing studies in sperm biology have revealed that the sperm cell contains a myriad of molecular constituents that may play a role post-fertilization, influencing the oocyte and embryo development (Figure 1). One such group of molecules are the small non-coding RNAs, which include the microRNAs (miRNAs).
MicroRNAs
MiRNAs are small non-coding single-strand RNA molecules with sizes ranging from 17-26 nucleotides. The first miRNA to be discovered was lin-4 in the nematode, Caenorhabditis elegans. Lin-4w miRNA was detected with conserved complementary site on lin-14a mRNA and subsequently affected the larvae developmental timing by targeting mRNA and inhibiting its function [1]. Since then, miRNAs have emerged to play key roles in many biological functions through binding to 3’UTR (untranslated region) of specific mRNAs leading to either mRNA degradation or protein translation inhibition [2]. These miRNAs are involved in regulating cell functions including gene expression during development, differentiation, cell proliferation and apoptosis [3,4].
In addition to having an influence in normal biological and developmental processes, miRNAs have been found to be involved in many pathogenic processes including Cardio Vascular Diseases (CVD), cancer, inflammatory or autoimmune diseases, metabolic disorders and neurodegenerative disorders [5].
MiRNA Biogenesis
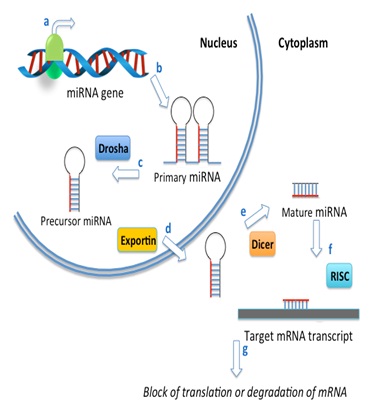
MiRNAs are encoded in the genome and transcribed by RNA polymerase II to form long primary miRNA transcripts (pri-miRNAs) that contain a cap structure at the 5′ end and are poly-adenylated at the 3′ end. The primary MiRNA sare subsequently processed in the nucleus by Drosha to form shorter stem loop structures called precursor-miRNAs or pre-miRNA (Figure 2). These pre-miRNAs are then transported to the cytoplasm by an Exportin-5 and processed by the RNase III enzyme called Dicer forming a short double-stranded miRNA duplex. This mature miRNA assembles into a protein-RNA complex called RISC (RNA Induced Silencing Complex) containing members of the Argonaut family. This multi component complex targets mRNA transcripts inhibiting gene expression by inhibiting impeding ribosomal attachment, or through degradation of the mRNA transcript [6-8].
Sperm-Specific MiRNAs and Male Infertility
The prevalence of infertility is estimated to be 15% globally, with males accounting to approximately 50% of cases. The causes of Male Factor Infertility (MFI) include infections, testicular injury, endocrinopathies and genetic disorders. However, a significant proportion of MFI is considered idiopathic, which is accompanied with low sperm numbers (oligozoospermia) (azoospermia, cryptozoospermia, and oligo-asthenozoospermia) and/or poor sperm motility (asthenozoospermia) (asthenozoospermia) [9].
Sperm contains a complex RNA population including messenger RNA (mRNA), transfer RNA (tRNA) and various non-coding RNAs (ncRNAs) such as the miRNAs [10]. The advancements in sequencing technology have enabled researchers to explore, in depth, the microRNA profile of many cell types [11] including sperm, where a plethora of novel sperm-specific miRNAs have been described [12,13]. Increased miRNA levels at the pachytene spermatocyte stage and the post-meiotic stage during spermatid development together with specific miRNAs expression in mouse test is suggesting that miRNAs have a role in regulating spermatogenesis [14-17].
Many reports have implicated dysregulation of sperm-specific miRNAs as likely to play an essential role in male fertility. In one study 154 differentially down regulated and 19 up-regulated miRNAs were found between a non-obstructive azoospermic group and a control group [18]. Additionally, it was noted that miRNA levels in seminal plasma revealed 19 miRNAs with altered expression in the seminal plasma and suggested 7 miRNAs that could be used as biomarkers of male infertility [19].
In a more recent study, it was found in a recent study that 50 miRNAs were up-regulated and 27 miRNAs down-regulated in asthenozoospermic males. In oligo-asthenozoospermic males (reduced sperm number and motility), 42 miRNAs were up-regulated and 44 miRNAs down-regulated when compared with normozoospermic males [20]. They showed miRNAs that exhibited the highest fold changes were miR-34b, miR-122, and miR-1973 in samples from asthenozoospermic men and miR-34b, miR-34b, miR-15b, miR-34c-5p, miR-122, miR-449a, miR-1973, miR-16, and miR-19a in samples from oligo-asthenozoospermic men. These data revealed a comprehensive number of miRNAs that were differentially expressed in asthenozoospermic and oligo-asthenozoospermic males compared with normozoospermic males, and provide support for the analysis of miRNA profiles as a future diagnosing tool for male infertility.
Salas-Huetos et al., another study, compared the miRNA expression profile of sperm from three different infertile populations and one fertile group of men. They demonstrated that sperm from patients with seminal alterations exhibit a differential miRNA profile. Evaluating 736 miRNAs in these sperm, they found that has-miR-34b-3p correlated with age, the has-miR-629-3p with sperm motility, and the has-miR-335-5p, has-miR-885-5p, and has-miR-152-3p with sperm concentration [21].
Recently, it was demonstrated that dysregulation of miRNAs (miR-29 and miR-424) can induce DNA double-strand breaks during spermatogenesis - further highlighting the increasing role miRNAs have on sperm integrity and function [22].
In addition to influencing spermatogenesis and sperm function-sperm-specific RNAs are introduced into the oocyte upon fertilization where it can contribute to gene regulation and embryo development [23].
Amanai et al., demonstrated that mature spermatozoa in mice contain a broad profile of miRNAs, and that potential mRNA targets of these miRNAs are expressed in metaphase II oocytes [24]. Early studies with human sperm revealed a unique population of sperm-specific miRNA, which are postulated to regulate sperm function [25] and may play an important role early zygotic and embryonic development [26].
Sperm MiRNA and Transgenerational Inheritance
Epigenetic Transgenerational Inheritance (ETI) is defined as germ line transmission of epigenetic information between generations, via gametes, in the absence of alterations in the DNA sequence, and epigenetic modifications, such as chromatin/histone modification, DNA methylation and acetylation can contribute to alter gene expression in heritable manner without affecting the underlying genomic sequences [27,28]. There is increasing development highlighting the role of ncRNAs in ETI and in particular the role of sperm-borne ncRNAs such as miRNAs [29].
MiRNAs are abundant in the mature sperm in mammals, and are introduced to the oocyte at fertilization, where they may transfer transgenerational inheritance [30]. There have been several interesting studies exploring the link between ETI (through transmission of sperm miRNAs) and environmental exposures. For example, paternal exposure to tobacco smoke and its components can alter the level of microRNAs in sperm [31]. Another study revealed that the predominant pathways mediated by miRNAs differentially expressed in the sperm of smokers were involved in cell proliferation, differentiation and death, pathways important during spermatogenesis and early embryo development. Thus, indicating potential epigenetic role for miRNAs in the multigenerational toxicity of cigarette smoke [32]. Diet can also alter miRNA expression and have transgenerational consequences as demonstrated in a study conducted by Fullston et al., (2013). This study revealed that diet-induced paternal obesity modulates sperm miRNA content, which initiate the transmission of obesity and impaired metabolic health to future generations [33]. Furthermore, a more recent study showed ETI through sperm-borne miRNA induced metabolic disorders in female offspring and with some intervention (diet and exercise) improved their metabolic health and prevents the metabolic syndrome in their female offspring [34].
Gapp et al., recently provided evidence demonstrating demonstrated that RNA-dependent processes contribute to the transmission of acquired traits in mammals. They carried out elaborate experiments and found that traumatic stress in early life altered mouse sperm miRNA expression, and behavioural and metabolic responses in the progeny. Injection of sperm miRNAs from traumatized males into fertilized wild-type oocytes reproduced the behavioural and metabolic alterations in the resulting offspring [35].
MiRNAs as a Biomarker for Male Infertility
Profiling miRNAs is important in understanding how they regulate different biological pathways and contribute to disease, including infertility. Reliable and reproducible research on miRNA function depends on sample isolation, amplification and analysis specifically designed for miRNAs. There are several companies supplying kits explicitly for miRNA isolation and identification. These kits enable the investigator to isolate high quality miRNA for downstream applications. Once isolated the miRNA is reverse transcribed, converting all of the miRNA in to cDNA. These cDNA libraries can be used as templates for quantitative (Real Time) Polymerase Chain Reaction (qPCR) to profile the miRNA [36].
MiRNAs can be isolated and detected in bodily fluids such as plasma, saliva, semen and vaginal fluid [37]. Therefore, utilizing this technology to explore the link between miRNA expression and male infertility can have great potential as anon-invasive biomarker for male infertility [38]. MiRNAs and other noncoding RNAs can be transported in the semen via seminal exosomes where they may be transferred to other cells with potential regulatory functions [39]. There are a growing numbers of studies reporting miRNA profiling in sperm form fertile and infertile males. One study described five miRNAs (has-miR-34b, has-miR-34b, has-miR-34c-5p, has-miR-429, and has-miR-122) using qRT-PCR analysis in sub fertile and non obstructive azoospermic patients, and control subjects. They reported that hsa-miR-429 was significantly increased and the four other miRNAs were decreased in both tested groups compared with normal control subjects. This study concluded that these five miRNAs have potential as novel non-invasive biomarkers to diagnose patients with sub fertility [40]. In another study the expression level of 736 miRNAs was evaluated. The investigators reported that up-regulated miRNAs presented an enriched localization in introns, affecting relevant genes for spermatogenesis and the predicted targets of the miRNAs contained critical genes associated to infertility [21]. MiRNAs can also be a predictor for sperm DNA damage, as demonstrated by Zhao et al., (2015), where they showed that miR-424/322 is involved in sperm DNA damage, and that the dysregulation of this miRNA can induce DNA double-strand breaks during spermatogenesis [22].
Human sperm contain a unique family of miRNA, which is responsible for normal spermatogenesis and sperm function and their expression profile will serve as a sensitive, selective and non-invasive diagnostic tests for male infertility.
Conclusion
The field of andrology has greatly expanded beyond the diagnostic field, merging with genetics, molecular biology and evolutionary biology. The sperm cell bestows to the oocyte much more than the paternal genome, centrioles and the activating factor - it provides the oocyte with a molecular signature that can influence not only early embryogenesis, but also determine the health of the progeny. MiRNAs have shown to be of immense importance in many cellular processes and we are only at the beginning of our understanding of their role in both health and disease.
Figures
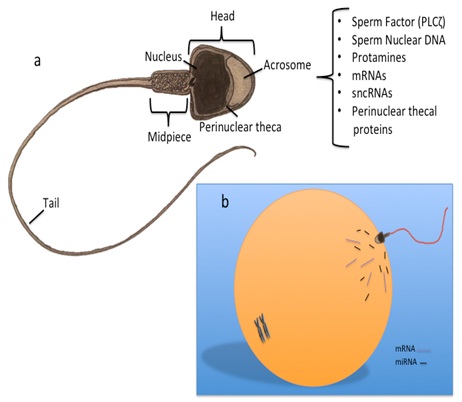
Figure 1: Sperm structure and constituents transferred to the oocyte at fertilization.
a) The fertilising sperm introduces more than the paternal genome, the oocyte-activating factor (PLCζ), proteins form post-acrosomal, and perinuclear thecal proteins and centrioles; b) Sperm also contain a complex population of RNA, including messenger RNA (mRNA) transcripts, small non-coding RNAs (sncRNA) and microRNAs (miRNA), which are also introduced in to the ooplasm at fertilization.
Figure 2: Micro RNA biogenesis.
(a) (a) Transcription of miRNAs is carried out by RNA polymerase II; b) leading to the generation of a primary miRNA transcript (pri-miRNA). These large primary transcripts are self-complementary and fold into a double-strand hairpin structure; c) The primary miRNA is endonucleolytically cleaved by an RNAse III superfamily member endonuclease called Drosha, resulting in the generation of smaller precursor molecules called precursor-miRNAs (pre-miRNA); d) Pre-miRNAs are then exported from the nucleus via exportin-5; e) After export from the nucleus to the cytoplasm, the pre-miRNA is further processed by Dicer to generate short, double-stranded miRNAs; f) These pre-miRNAs interact with Argonaute (AGO) and other regulatory proteins forming a miRNA-protein complex called RISC (RNA-induced silencing complex) where they are converted to mature, single-stranded miRNAs; and g) Finally, mature miRNAs direct RISC to target mRNA, resulting in inhibition of gene expression by perturbation of ribosomal RNA binding to the mRNA transcript or through mRNA degradation.