
Effect of Genetic Factors on Atopic and Non Atopic Asthmatic and Allergic Rhinitis Saudi Children in Taif Area
*Corresponding Author(s):
Yousri M HusseinDepartment Of Medical Biochemistry, Zagazig University, Zagazig, Egypt
Tel:+966 201001752426,
Email:yousrihussein@hotmail.com
Abstract
To assess the value of serum Interleukin-13 (IL-13) levels as an immunological marker in atopic upper respiratory diseases, to clarify its differences in atopic and non atopic bronchial asthma and to determine the role of an Interleukin -13 receptor alpha 1 (IL-13 Rα1) gene Single Nucleotide Polymorphism (SNP) (A1398G) in the pathogenesis of these diseases.
Methods
Seventy-five patients were compared with 25 age-matched healthy volunteers. Serum total immunoglobulin E (Ig E) and IL-13 levels were measured by enzyme-linked immune sorbent assay and the IL-13Rα1 gene (A1398 G) was screened by specific polymerase chain reaction.
Results
There was a non significant association between G allele frequencies of the IL-13Rα1 (1398) gene polymorphism (42%, 38% and 30% for atopic asthma, non atopic asthma, and allergic rhinitis, respectively) as compared to in controls. There were a significant increase in the serum level of total IgE & IL-13 towards heterozygous AG and homozygous GG than homozygous AA in atopic asthma, non atopic asthma, and allergic rhinitis patients. There was a significant increase in the serum level of total IgE & IL-13 towards homozygous GG than heterozygous AG in atopic asthma (p=0.035), non atopic asthma (p=0.014), and allergic rhinitis patients (p=0.003) for IgE and (p<0.001) in all groups for IL-13 as shown by LSD test.
Conclusion
Serum interleukin (IL) 13 can be used as an immunological marker in atopic upper respiratory diseases and to differentiate between atopic and non atopic bronchial asthma.
Keywords
INTRODUCTION
Asthma is associated with atopy and with IgE-mediated inflammation of the airways, which occurs via release of Interleukin (IL)-4 and IL-13, both of which have been included in the pathogenesis of asthma in multiple human and animal studies [1]. Human IL-13 is a protein with a molecular mass of 13 KDA, which has four α helical bundles (αA, αB, αC and αD) [2]. In Saudi Arabia, the prevalence of asthma is higher than in other Arab countries, with substantial regional variations [3-5]. Saudi Arabia has increasing prevalence of childhood atopic asthma. Allergic disease was reported to be very common in primary school-aged children in different areas of Saudi Arabia, including Taif city, with figures closer to the highest risk regions in the world [6-9].
It has an important role as an effector molecule in asthma through multiple mechanisms, including induction of production of immunoglobulin (IgE) by B-cells [10], attraction of eosinophils to the airway [11], metaplasia of goblet cell and increase mucus secretion, and airway remodeling [12]. A wealth of data supports a role for IL-13 in mediating asthma pathology. IL-13 produces its effect via receptor which is heterodimeric and composed of two membrane proteins, [13] IL-4Rα and either a low affinity IL-13Rα1 or high affinity IL-13Rα2. [14] IL-13 binds to IL-13 binding chain (IL-13Rα1) at low affinity in the absence of IL-4Rα, whereas in the presence of IL-4R, the site becomes of high affinity [15]. IL-13Rα1 is widely expressed and has been detected on nearly every cell tested except human T cells, while, the human IL-13Rα1 gene is present on the X q24 chromosome possibly suggesting a role in X-linked immune disease [16].
The C-terminal alpha helix D, which is one of the four alpha helices that constitutes IL-13, contains key residues for binding with both IL-13Rα1 and IL-13Rα2, whereas IL-13 interaction with IL-4Rα is mediated via helices A and C [3]. Analysis of crystal structures of ternary complexes composed of IL-13 or IL-4 binding with IL-13Rα1 and IL 4Rα chain gives idea about shared receptor interactions with distinct cytokine, explaining the different affinities of cellular responses to IL-4 and IL-13[16] Association of the IL-13Rα1/IL-4R receptor complex by IL-13 results in stimulation of a variety of signal transduction pathways. Upon forming a dimmer of IL-13Rα1 with IL-4Rα, JAK1 and Tyk-2 kinases become phosphorylated and activated which lead to phosphorylation of tyrosine residues on the IL-4Rα and IL-13Rα1 chains [17].
Polymorphisms in these receptor molecules contribute to the genetic effects on asthma susceptibility and atopic diseases as allergic rhinitis. For example, asthma susceptibility was significantly elevated in Korean children by gene–gene interaction between IL-4 T-590C and IL-4Rα Gln 551 Arg alleles [18-19] and between IL-13 C-1112T and IL- 4Rα Ser478 Pro alleles in Dutch population [20]. Also, a non coding polymorphism in IL-13Rα1 (A+1398G) was linked to increased IgE levels in British population [21].
On the basis of the important role of the IL-13/IL-4 pathway in atopy and asthma, it was hypothesized that genetic variation in IL-13Rα1 may lead to the development and/or predict severity of asthma and atopy [22-24].
PATIENTS AND METHODS
Participants
Collection of blood samples
Determination of serum IL-13
Total IgE measurements
Detection of (A1398G) gene polymorphism
The primer sequences used for the IL-13Rα1 (1398) gene polymorphism were as follows:
5' – TCA GTC ATG GAG ATA ATT TA 3' (sense)
5' – TGA GCT GCC TGT TTA TAA AT 3' (antisense)
The products were digested using MseI (New England Biolabs), which digested the +1398A allele into 85 and 45 bp fragments and yielded a single 130 bp band for the +1398G allele. The 20 μl of PCR products were digested with 5 U of the restriction enzyme at 65ºC for 16 h, and separated on 3% agarose gel stained with ethidium bromide.
Data analysis
RESULTS
(A1398G) Frequencies
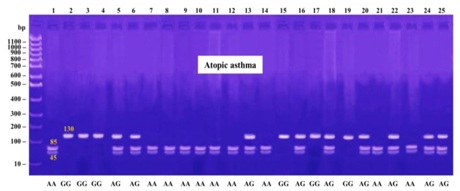
1398G allele frequencies
Association between (A1398G) polymorphism and IL-13 and IgE Levels
IL-13 and IgE levels
| Control Group | Atopic Asthma Group | Non Atopic Asthma Group | Allergic Rhinitis Group | |
| No Percent | No Percent | No Percent | No Percent | |
| AA | 13 52% | 10 40% | 11 44% | 14 56% |
| AG | 8 32% | 9 36% | 9 36% | 7 28% |
| GG | 16 4% | 24 6% | 5 20% | 4 16% |
| X2 | 3.4 | 1.35 | 0.41 | |
| p | 0.182(>0.05) | 0.5 (>0.05) | 0.812(>0.05) |
| Group | No percent | Odds Ratio (95% CI) |
| Control group | 16 32% | |
| Atopic asthma | 21 42% | 1.5 (0.8-2.7) |
| Non atopic asthma | 19 38% | 1.3 (0.7-2.3) |
| Allergic rhinitis | 15 30% | 0.9 (0.5-1.65) |
| Groups | Measurement | AA | AG | GG | Pbvalue |
| Atopic asthma | Serum Il-13 pg/ml | 13.73 ± 0.46 | 15.98 ± 0.5 | 18.87 ± 1.06 | <0.001 |
| Serum IgE IU/ml | 244.8 ± 42.6 | 360.66 ±10.67 | 394.3 ± 5.83 | <0.001 | |
| Nonatopic asthma | Serum Il-13 pg/ml | 4.37 ± 0.54 | 6.38 ± 0.69 | 7.88 ± 0.21 | <0.001 |
| Serum IgE IU/ml | 68.63 ± 9.99 | 99.66 ± 6.65 | 111.4 ± 2.19 | <0.001 | |
| Allergic rhinitis | Serum Il-13 pg/ml | 14.12 ± 0.82 | 16.82 ± 0.69 | 19.26 ± 1.0 | <0.001 |
| Serum IgE IU/ml | 255.2 ± 27.27 | 337 ± 14.47 | 384.5 ± 12.44 | <0.001 |
| Measurement | Control | Atopic asthma | Non atopic asthma | Allergic rhinitis | |
| IgE IU/ml | 70.1 ± 18.5 | 322.32 ± 71.126* | 88.36 ± 19.84 | 298.8 ± 56.85* | |
| Il-13 pg/ml | 5.828 ± 1.2 | 15.78 ± 2.1* | 5.79 ± 1.5 | 15.7 ± 2.1* | |
DISCUSSION
However, IL-13 responsiveness has been reported in the absence of IL-4Rα, suggesting the presence of alternate receptor forms. AHR, airway mucus production, and lung eosinophilia were distinguished in mice with transferred OVA-specific IL-13 producing T cells but lacking IL-4Rα, but not in those lacking STAT6 [32]. Also, in mice lacking IL-13 airway inflammation, fibrosis, and mucus cell hyperplasia, were diminished [33], but persisted in animals deficient in IL-4 [34] or IL-4Rα [33], suggesting that IL-13 may produce its effects through an IL-4Rα-independent pathway in this asthma model [15-24,33-35].
Three fibronectin type III sub units (D1, D2, and D3) compose the extracellular r portions of IL-13Rα1[36]. Significant impairment of the IL-13 response may be due to mutation of IL-13 and IL-13Rα, as found in mutation of Leu 319 and Tyr 321 in the D2 and D3 respectively of IL-13Rα [36]. IL-13 induced signals or responses was not able to be mediated through IL-13Rα1 lacking the intracellular domain, supporting the possibility that IL-13Rα1 is required for signaling [37].
We reported that there was a no significant association between IL-13Rα1 polymorphism (+A1398G) and the susceptibility of atopic asthma, non atopic asthma or allergic rhinitis in Egyptian children while hetero- or homozygosity for the risk allele of IL-13Rα1 A+1398G was significantly associated with increased total IgE & IL-13 levels in children with atopic asthma, non atopic asthma and allergic rhinitis. Konstantinidis et al., [38] reported that no association between IL-13Rα1 (A+1398G) and asthma susceptibilityor severity or with the development of atopic phenotype in Caucasian families while, IL-13Rα1 polymorphism (A+1398G) included in the control of IgE production.
Kim et al., [39] found that IL-13Rα1 (+A1398G) were not linked to the susceptibility of asthma or atopic asthma and that hetero- or homozygosity for the risk allele of IL-13Rα1 A+1398G was significantly associated with increased total IgE levels in children with atopic asthma, non atopic asthma and allergic rhinitis. Heinzmann et al., [21] reported that non coding polymorphism in IL-13Rα1 (A+1398G) was associated with increased level of IgE in British population with restriction among different ethnic groups as in Japanese people. This may suggest that this non-coding polymorphism of IL-13Rα1 has a functional effect for the binding of IL-13, or alternatively that the IL-13Rα1 A+1398G polymorphism is associated with as yet undiscovered polymorphisms in the regulatory or codingregions of the gene encoding IL-13Rα1. Another possibility is that IL-13Rα1 could have additional unknown signaling functions that are impacted by this polymorphism.
Collectively, these studies indicate that IL-13Rα1 is likely to play a critical role not only in binding but also in signaling of IL-13. We reported also a highly significant increase of serum total IgE & IL-13 levels in atopic asthma and allergic rhinitis groups as compared with the control group and in atopic asthma and allergic rhinitis when compared with non atopic asthma. There was no statistically significant difference between non atopic asthma and control group or between atopic asthma and allergic rhinitis groups and there is a positive correlation between IL-13 and IgE levels in these groups. This finding consistent with observations by EL-Helaly et al [40], Turato et al., [41] and Gergen et al.,[42] who reported from National health and nutrition examination survey (2005, 2006) that total IgE level predicted asthma among atopic subjects but not among non atopic individuals. Elevation of serum IL-13 in asthma group agreed with Lee et al., [43] who evidenced that the expression of IL13, IL4, and IL5 were increased in acute asthmatic patients so they may be deeply involved in the pathogenic process of asthma [44].
Yang et al., [45] suggested that anti-IL-13 monoclonal antibody inhibits airway hyper responsiveness, inflammation and airway remodeling in a chronic mouse model of asthma. These findings also confirmed with Kumar et al., [33] and Follettie et al., [46] who concluded that inhibition of IL-13 has considerable potential as a therapeutic strategy in chronic asthma. Thom et al., [47] optimized the affinity of a human IL-13-neutralizing antibody, a therapeutic candidate for the treatment of asthma, more than 150-fold. Ippoliti et al., [48] [who observed a significant reduction in asthma and rhinitis scores in the immunotherapy group compared with the placebo group associated with a significant decrease in IL-13 after 6 months of therapy.
We can explain the non significant difference in IL-13 level between atopic asthma and allergic rhinitis that both conditions have similar mechanisms and underlying pathogenesis, many of the cells, mediators, cytokines, and neurotransmitters involved in the biology of asthma and rhinitis are the same [22]. Indeed, up to 45% of patients with asthma have allergic rhinitis while 93.5% of those with allergic rhinitis were also asthmatic [49]. Feleszko et al., [50] found a positive correlation between IL-13 levels and serum IgE concentrations in children with allergic asthma. This can be explained by that the most important inducers of the release of IgE are IL-4 and IL-13 [51-54]. These cytokines stimulate transcription of the gene for the epsilon class of the constant region (C€) of the immunoglobulin heavy chain [22,51,55-56]. In the other hand, Hussein et al., [57] denied this correlation suggesting that IL-13 role in the pathogenesis of allergic diseases was not only by inducing IgE production but also by other mechanisms. This hypothesis was concordant with animal models where mechanisms responsible for asthma and allergic rhinitis were independent of IgE levels [58] .
However, other studies doubt on the role of serum total IgE as an important indicator of atopic diseases [59,60] found that both atopic and non atopic asthmatics had raised serum total IgE levels. The difference may be due to different population who were included in the study or to the different age groups or due to the severity of the disease. Future work will be required to identify gene–gene and gene–environment interactions on a genome-wide level, with the aim of fully understanding the genetic risk factors for asthma and atopy, the pathogenesis of these common diseases, and perhaps describing new treatment strategies aimed at changing IL-13 signaling via IL-4Rα/IL-13Rα1.
CONCLUSION
ACKNOWLEDGMENT
REFERENCES
- Floistrup H, Swartz J, Bergstrom A, Alm JS, Scheynius A, et al. (2006) Allergic disease and sensitization in Steiner school children. J Allergy Clin Immunol 117: 59-66.
- Moy FJ, Diblasio E, Wilhelm J, Powers R (2001) Solution structure of human IL-13 and implication for receptor binding. JMB 310: 219-230.
- Al-Dawood K (2000) Epidemiology of bronchial asthma among schoolboys in Al-Khobar city, Saudi Arabia: cross-sectional study. Croat Med J 41: 437-441.
- Al-Frayh AR, Hasnain SM (2007) Prevalence of bronchial asthma in children in Saudi Arabia. World Allergy Organization Journal 167-168.
- Al Frayh AR, Shakoor Z, Gad El Rab MO, Hasnain SM (2001) Increased prevalence of asthma in Saudi Arabia. Ann Allergy, Asthma Immunol? 86: 292-296.
- Al-Makoshi A, Al-Frayh A, Turner S, Devereux G (2013) Breastfeeding practice and its association with respiratory symptoms and atopic disease in 1-3-year-old children in the city of Riyadh, central Saudi Arabia. Breast Feed Med 8: 127-133.
- Alshehri MA, Abolfotouh MA, Sadeg A, Al Najjar YM, Asindi AA, et al. (2000) Screening for asthma and associated risk factors among urban school boys in Abha city. Saudi Med J 21: 1048-1053.
- Sabry EY (2011) Prevalence of allergic diseases in a sample of Taif citizens assessed by an original Arabic questionnaire (phase I) A pioneer study in Saudi Arabia. Allergol immunopathol 39: 96-105.
- Nahhas M, Bhopal R, Anandan C, Elton R, Sheikh A (2012) Prevalence of allergic disorders among primary school-aged children in Madinah, Saudi Arabia: two-stage cross-sectional survey. PLoS One 7: 36848.
- Defrance T, Carayon P, Billian G, Guillemot JC, Minty A, et al. (1994) Interleukin 13 is a B cell stimulating factor. J Exp Med 179: 135-143.
- Wardlaw AJ (2001) Eosinophil trafficking in asthma. Clin Med (Lond) 1: 214-218.
- Richter A, Puddicombe SM, Lordan JL, Bucchieri F, Wilson SJ, et al. (2001) The contribution of interleukin (IL)-4 and IL-13 to the epithelial-mesenchymal trophic unit in asthma. Am j Respir Cell and Mol Biol 25:385-391.
- Madhankumar AB, Mintz A, Debinski W (2002) Alanine-scanning mutagenesis of alpha-helix D segment of interleukin-13 reveals new functionally important residues of the cytokine. J Biol Chem 277: 43194-43205.
- Akdis CA, Akdis M (2009) Mechanisms and treatment of allergic disease in the big picture of regulatory T cells. J Allergy Clin Immunol 123: 735-746.
- LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, et al. (2008) Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132: 259-272.
- Hussein YM, Ahmad AS, Ibrahem MM, Elsherbeny HM, Shalaby SM, et al. (2011) Interleukin 13 receptors as biochemical markers in atopic patients. J Investig Allergol Clin Immunol 21: 101-107.
- Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE (1999) The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol 17: 701-738.
- Lee SL, Wong W, Lau YL (2004) Increasing prevalence of allergic rhinitis but not asthma among children in Hong Kong from 1995 to 2001 (Phase 3 International Study of Asthma and Allergies in Childhood). Pediatr Allergy Immunol 15: 72-78.
- Hussein YM, Awad HA, Shalaby SM, Ali AS, Alzahrani SS (2012) Toll-like receptor 2 and Toll-like receptor 4 polymorphisms and susceptibility to asthma and allergic rhinitis: A case–control analysis. Cell Immunol 274: 34-38.
- Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, et al. (2002) Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet 70: 230-236.
- Heinzmann A, Mao XQ, Akaiwa M, Kreomer RT, Gao PS, et al. (2000) Genetic variants of IL-13 signalling and human asthma and atopy. Hum Mol Genet 9: 549-559.
- Hussein YM, El-Tarhouny SA, Shalaby SM, Mohamed RH, Hassan TH, et al. (2011) Interleukin-13 receptor A1 gene polymorphism and IL-13 serum level in atopic and non-atopic Egyptian children. Immunol invest 40: 523-534.
- Hussein Y, Allah SA, Mahmoud S, Ahmed A (2006) Impact of IL-13 gene mutations in atopic diseases. EJBMB 24: 123-127.
- Hussein Y, Abou El YM, Mahmoud Y, Mahmed H, Rasha L (2006) Some biochemical markers in atopic patients and PCR-based assay for detection of R576 IL-4 receptors allele gene. Zagazig University Medical Journal 12: 4307-4321.
- Meltzer EO (1988) Evaluating rhinitis: clinical, rhinomanometric, and cytologic assessments. J Allergy Clin Immunol 82: 900-908.
- [No authors listed] (1995) Standardization of Spirometry, 1994 Update. American Thoracic Society. Am J Respir Crit Care Med 152: 1107-1136.
- Kirkwood B, sterne AJC (1989) Essentials of Medical Statistics, (2nd edn). Blackwell Scientific Publication, London, England.
- Sengler C, Lau S, Wahn U, Nickel R (2002) Interactions between genes and environmental factors in asthma and atopy: new developments. Respir res 3: 7.
- Vercelli S, Ferriero G, Sartorio F, Stissi V, Franchignoni F (2009) How to assess postsurgical scars: a review of outcome measures. Disabil Rehabil 31: 2055-2063.
- Mobasher A, El-Shahat H, Gabry M, Affara N, Hussein Y (2004) Eosinophilic Cationic Protein (ECP) in bronchial asthma, Role of treatment. Egypt J Chest Dis & Tub 53: 17-26.
- Andrews AL, Holloway JW, Puddicombe SM, Holgate ST, Davies DE (2002) Kinetic analysis of the interleukin-13 receptor complex. J Biol Chemistry 277: 46073-46078.
- Mattes J, Yang M, Siqueira A, Clark K, MacKenzie J, et al. (2001) IL-13 induces airways hyperreactivity independently of the IL-4R alpha chain in the allergic lung. J Immunol 167: 1683-1692.
- Kumar RK, Herbert C, Yang M, Koskinen AML, McKenzie ANJ, et al. (2002) Role of interleukin-13 in eosinophil accumulation and airway remodelling in a mouse model of chronic asthma. Clin Exp Allergy 32: 1104-1111.
- Foster PS, Ming Y, Matthei KI, Young IG, Temelkovski J, et al. (2000) Dissociation of inflammatory and epithelial responses in a murine model of chronic asthma. Lab Invest 80: 655-662.
- Hussein YM, Shalaby SM, Nassar A, Alzahrani SS, Alharbi AS, et al. (2014) Association between genes encoding components of the IL-4/IL-4 receptor pathway and dermatitis in children. Gene 545: 276-281.
- Arima K, Sato K, Tanaka G, Kanaji S, Terada T, et al. (2005) Characterization of the interaction between interleukin-13 and interleukin-13 receptors. J Biol Chem 280: 24915-24922.
- Orchansky PL, Ayres SD, Hilton DJ, Schrader JW (2005) An interleukin (IL)-13 receptor lacking the cytoplasmic domain fails to transduce IL-13-induced signals and inhibits responses to IL-4. J Biol Chem 272: 22940-22947.
- Konstantinidis AK, Barton SJ, Sayers I, Yang IA, Lordan JL, et al. (2007) Genetic association studies of interleukin-13 receptor alpha1 subunit gene polymorphisms in asthma and atopy. Eur Respir 30: 40-47.
- Kim H-B, Lee Y-C, Lee S-Y, Jung J, Jin H-S, Kim (2006) Gene-gene interaction between IL-13 and IL-13Ralpha1 is associated with total IgE in Korean children with atopic asthma. Journal of Human Genetics 51: 1055-1062.
- El-Helaly N, El-Wan A, Kamel Y, Nabih M, Mahmoud H (2009) Eosinophil-Derived Neurotoxin Versus Immunoglobulin E as Biomarkers for Evaluation of Bronchial Asthma. J Biol Chem 9: 165-169.
- Gergen PJ, Arbes SJ Jr, Calatroni A, Mitchell HE, Zeldin DC (2009) Total IgE levels and asthma prevalence in the US population: Results from the National Health and Nutrition Examination Survey 2005-2006. J Allergy Clin Immunol 124: 447-453.
- Lee YC, Lee KH, Lee HB, Rhee YK (2001) Serum levels of interleukins (IL)-4, IL-5, IL-13, and interferon-gamma in acute asthma. J Asthma 38: 665-671.
- Abdel-Mawla MY, Mostafa Y, Abuel-Majd Y, Attwa R (2009) Detection of R576 interleukin-4 receptor an allele gene, serum interleukin-4, and eosinophilic cationic protein in atopic dermatitis patients. Indian J Dermatol 54: 31-35.
- Yang G, Volk A, Petley T, Emmell E, Giles-Komar J, et al. (2004) Anti-IL-13 monoclonal antibody inhibits airway hyperresponsiveness, inflammation and airway remodeling. Cytokine 28: 224-232.
- Follettie MT, Ellis DK, Donaldson DD, Hill AA, Diesl V, et al. (2006) Gene expression analysis in a murine model of allergic asthma reveals overlapping disease and therapy dependent pathways in the lung. Pharmacogenomics J 6: 141-152.
- Thom G, Cockroft AC, Buchanan AG, Candotti CJ, Cohen ES, et al. (2006) Probing a protein-protein interaction by in vitro evolution. Proceedings of the National Academy of Sciences of the United States of America 103: 7619-7624.
- Ippoliti F, De Santis W, Volterrani A, Lenti L, Canitano N, et al. (2003) Immunomodulation during sublingual therapy in allergic children. Pediatr Allergy Immu 14: 216-221.
- Cengizlier MR, Misirlioglu ED (2006) Evaluation of risk factors in patients diagnosed with bronchial asthma. Allergol Immunopathol (Madr) 34: 4-9.
- Feleszko W, Zawadzka-Krajewska A, Matysiak K, Lewandowska D, Peradzy?ska J, et al. (2006) Parental tobacco smoking is associated with augmented IL-13 secretion in children with allergic asthma. J Allergy Clin Immunol 117: 97-102.
- Hussein YM, Alzahrani SS, Alharthi AA, Ghonaim MM, Alhazmi AS, et al. (2014) et al. Association of serum cytokines levels, interleukin 10 −1082G/A and interferon-γ +874T/A polymorphisms with atopic asthma children from Saudi Arabia. Cell Immunol 289: 21-26.
- Hussein YM, Shalaby SM, Zidan HE, Sabbah NA, Karam NA, et al. (2013) CD14 tobacco gene-environment interaction in atopic children. Cell Immunol 285: 31-37.
- Hussein YM, Alzahrani SS, Alharthi AA, Alhazmi AS, Ghonaim MM, et al. (2016) Gene Polymorphism of Interleukin-4, Interleukin-4 Receptor and STAT6 in Children with Atopic Dermatitis in Taif, Saudi Arabia. Immunol Invest 45: 223-234.
- Zahran F, Hussein YM, Ashour E, Sayed M, Shalaby SM, et al. (2013) Interleukin-4 and interleukin-4 receptor alpha polymorphisms in atopic dermatitis: A case-control study. BioChemistry: An Indian Journal 7: 89-96.
- Marinkovich VA (2001) Allergy and IgE antibodies. N Engl J Med 344: 1332-1333.
- Hussein PY, Zahran F, Ashour Wahba A, Ahmad AS, Ibrahiem MM, et al. (2010) Interleukin 10 receptor alpha subunit (IL-10RA) gene polymorphism and IL-10 serum levels in Egyptian atopic patients. J Investig Allergol Clin Immunol 20: 20-26.
- Hussein YM, Zahran F, El-Zaher AA, El Tarhouny SA, Shalaby SM (2012) Interleukin-10 gene polymorphism and its blood level as biochemical markers among Egyptian atopic patients. J Cell Sci Ther 3: 8.
- Hussein YM, El-Tarhouny SA, Shalaby SM, Mohamed RH, Hassan TH, et al. (2011) Interleukin-13 Receptor A1 Gene Polymorphism and IL-13 Serum Level in Atopic and Non-atopic Egyptian Children. Immunological Investigations 40: 523-534.
- Aliprantis AO, Wang J, Fathman JW, Lemaire R, Dorfman DM, et al. (2007) Transcription factor T-bet regulates skin sclerosis through its function in innate immunity and via IL-13. Proc Natl Acad Sci USA 104: 2827-2830.
- Bettiol J, Bartsch P, Louis R, De Groote D, Gevaerts Y, et al. (2000) Cytokine production from peripheral whole blood in atopic and nonatopic asthmatics: relationship with blood and sputum eosinophilia and serum IgE levels. Allergy 55: 1134-1141.
Citation: Hussein YM, Alghamdy AAN, Dahlawi HA, Zaini RG, Almaleki AA, et al. (2017) Effect of Genetic Factors on Atopic and Non Atopic Asthmatic and Allergic Rhinitis Saudi Children in Taif Area. J Allergy Disord Ther 3: 007.
Copyright: © 2017 Yousri M Hussein, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

