
Anti-Cancer Effects of Huaier on Prostate Cancer; miRNA-Mediated Transcription Control Induced Both Inhibition of Active Progression and Prevention of Relapse
*Corresponding Author(s):
Manami TanakaBradeion Institute Of Medical Sciences, Co. Ltd., Kanagawa, Japan
Tel:+81 463580952,
Email:tubu0125@gmail.com
Abstract
Prostate cancer is classified as mild malignant tumor, since the growth is slow, and also the early detection and monitoring of PSA and other cancer cell-specific markers are available among the target population in walk-in clinic even effect the bones and lymph nodes, it requires long term treatments which significantly decrease the lifespan in patients. Trametes robiniophila Murr (Huaier) has proved broad spectrum anti-cancer effects, which initiates the recovery of damaged bio physiological functions in the end by dose-dependent manner which proved the molecular basis of Huaier effects by total RNA and small non-coding RNA sequencing (“genome-scope” project) and that is based on the rescue of the disrupted transcriptional control based on individual capability and flexibility of genomic potential. Here we focused to show MEGA-DATA genome analysis results on inhibition of active progression in cancer in situ, and also prevents relapse of prostate cancer. Total sequencing of RNAs revealed massive SNP variances (average 89,473SNP variances per individual), however, it is unlikely that specific type variants influenced to the malignancy, process, and prognosis. On the other hand, the significant up-regulation and alteration of major ontogenesis and tumor suppressor genes were detected in transcribed genes, especially in altered (normalized) NFkB, TGFb, BRCA2 and p53 genes and their leading signaling pathways. These genetic alterations in transcriptomes were based on miRNA-mediated transcriptional control as reported drugs have not shown any effect. Thus, the present study provides the safe and effective treatment to prevent and inhibit prostate cancer progression, and also to maintain homeostasis in a long-range of stressful human life without excessive medical treatments.
Keywords
Cancer therapy; Genome-scope project; Huaier (Trametes robiniophila murr); miRNA-mediated transcription control; Prostate cancer
Introduction
Prostate cancer was reported as second-most effecting in the world, and estimated to have 1.2 million patients and 359,000 deaths in 2018 [1,2]. The incidence ratio and the numbers of deaths caused by prostate cancer are widely varied among countries, chiefly because of screening system in each nation [3].
In Japan, blood PSA screening together with MRI (magnetic resonance imaging) are used for 55 years elderly male population, which suggests about 50% population, has prostate cancer in situ http://www.zenritsusen-care.com/part02/index.html. However, at the same time, the mean period was estimated to be more than 40 years, which directed serious questions to radical treatments which significantly lower Quality of Life (QOL), often observed after surgical dissection. The two main complications encountered after prostatectomy and prostate radiotherapy are erectile dysfunction and urinary incontinence, mainly stress-type. Consequently, recent diagnostic system for prostate cancer is being innovated through accumulating experiences both from laboratory data and clinical significance for precise diagnosis and choice of treatment methods [4].
As for cancer-specific blood test, we have developed a novel technique by the detection of less than ng/ml blood level of cancer cell-specific molecule; Septin4/Bradeion [5-8]. Together with PSA detection, even blood test could provide the precise evaluation of prostate cancer only in active proliferating stage. Since the results from biological and biochemical investigations were obtained by using in vitro cultured cells chemical mutagenized, the results were often wide-split from the reality detected by clinical observations with advances in MRI technology [9,10], it is so important to distinguish and diagnose active cancer stage from latent cancer in situ to avoid unnecessary medical measures rather than regular monitoring tests for the life time long. In addition, it is more important to find the onset of activation of cancer in situ by stress accumulation.
Here we present the possible prevention of prostate cancer by the clinical observation of several cases at high risk of prostate cancer in situ, and how the efficacy of Huaier [11] appeared in the ancient Chinese literature from B.C. 247. Imperial expeditions in search for possible natural source to obtain immortality. However, successful discovery of Huaier could not provide the desired immortality, and at B.C. 220, the significant efficacy on health maintenance and longevity was reported, and that in dose-dependent manner. However, Trametes robiniophila Murr is very rare to find as natural source, until Chinese technology succeeded to cultivate and make a stable supply in quantity [12]. There are many kinds of Trametes species, for example Trametes versicolor (Krestin) has failed to induce any significant anti-cancer effects on colorectal cancer [13]. At present, only Huaier granule demonstrated its significant medical efficacy chiefly on many cancers as well as immunological disorders, skin problems, liver dysfunction and etc., and more importantly without any side effects or toxicity [11].
We have reported with simple and successful evidence that the rescue of Hippo signaling pathway was a molecular basis of anti-cancer effects of Huaier [14] in 2017. Thereafter many researches followed for the confirmation, to provide the evidence in vitro that deregulation in Hippo signaling pathway, disruption of transcription control is the major course of carcinogenesis [15-22]. However, there have not been appeared the additional successful controlling materials which contributes for cancer recovery so far.
Therefore, we initiated the clinical research to thorough understanding of molecular basis of Huaier effects by total RNA sequencing and also small non-coding RNA sequencing by total blood from the patients as samples. In China, it has become the trends to identify molecular basis of diseases by total understanding of the genomic and genetic information (MGA-DATA analysis) [23,24]. Our research to find the key molecules and pathways for cancer recovery, on the contrary to the results for carcinogenesis, shown surprising results beyond our expectations [25], results clearly indicated that the Huaier therapy influenced multi-events among whole genome-wide, and also in every process in transcription and translation to maintain cellular functions, reproduce cells and tissues as damage repair after cancer cell death and elimination of debris., i.e., for homeostasis thus named our project as “Genome-Scope”, to emphasize the importance of sky-high point of overview, and for the pattern recognition of genomic and genetic aspects required for the maintenance of homeostasis, not to be trapped by each trifle molecule or factor.
As reported, cancer patients who had enough genomic potential to tolerate total alterations by Huaier administration restored normal homeostasis [25], which resulted in successful cancer recovery. In prostate cancer, such a drastic genomic potential seemed to be unnecessary for the inhibition of cancer progression, and to keep cancer in situ in silent and inactive condition. However, minimum essential alterations were detected in major key molecules (NFkB, TGFb, BRCA2 and p53) [25-27]. We confirmed the rescue of transcription control in Hippo signalling pathway in the present study, but the complete rescue was observed among almost all the transcription control systems with the massive alterations in transcriptional factors (mean 1,115/person).These alterations were based on miRNA-mediated transcriptional control, and less part was supported by piRNA modifications, as predicted before [25].
Thus, the data obtained here could provide the actual solutions to the inhibition of cancer progression, the prevention of relapse on prostate cancer. The effects are observed in a dose and duration dependent manner, and that without any toxicity, side effects, and resistance in long use, since all the effects are originated as spontaneous reaction based on the individual genomic potential. In Japan, there are increasing male population with 3g/day Huaier administration for prostate cancer in situ, just as the same as the cases of liver dysfunction, and atopic dermatitis, and also for the prevention of serious progression in case of COVID-19 [25]. Huaier effects on prostate cancer provide its minimum-essential mode of action to control the disease without decreasing the QOL of the patients.
Materials And Methods
Project design and patients’ characterization
Huaier compounds were provided by the manufacturer following guidelines in good condition for maintenance, and provision to the volunteers. The patients were prescribed 20g per day of Huaier for 3 months, and suggested not to use other. The profiles and medical characteristics of thepatients were summarized in table 1. Patient No.1 has not shown any cancer biopsy. The present study was specifically focused on prostate cancer patients in this clinical research, which was strictly conducted according to the guidelines of the Declaration of Helsinki and the principles of good clinical practice.Written informed consent was obtained from the patients.This clinical research was applied according to the Consolidated Standards of Clinical Research Trials guidelines and Japanese Medical Association on 9th February 2018, and approved on 5th March, 2018 (ID: JMA-IIA00335).The project has been strictly conducted with a monthly review by the ethics committee with experts on Medicine, Nursing, Laws, Pharmaceutics and Business Community (first committee held on 9th February, 2018) [25]. The patient samples used in the present study were diagnosed chiefly by biopsy, together with blood PSA and Septin4/Bradeion tests. All patients underwent MRI examination regularly, once in 1-2 years after 50 years old.They are still having Huaier compound as their own choice, but the quantity varies among individuals (3 – 20 g per day).Periodical assessments of QOL and clinical tests are also being monitored at present.
|
Patient No. |
1 |
2 |
3 |
4 |
5 |
6 |
|
Age |
73 |
73 |
60 |
61 |
81 |
67 |
|
PSA |
17 |
within normal limits |
within normal limits |
0 |
beyond measurement limit |
within normal limits |
|
sepin 4 (Bradeion) |
within normal limits |
within normal limits |
within normal limits |
within normal limits |
within normal limits |
not examined |
|
diagnosis |
hyperplasia |
prostate cancer in situ (biopsy) |
prostate cancer in situ (biopsy) |
prostate cancer |
prostate cancer |
healthy control |
|
stage |
hyperplasia |
0 |
0 |
1 |
IV |
none |
|
metastasis |
|
|
|
|
lung, liver. Lymph nodes |
|
|
blood sampling time |
|
|
|
|
|
|
|
before Huaier administration |
O |
O |
O |
O |
O |
O* |
|
one month |
O |
O |
O |
O |
O |
O |
|
3 months |
O |
O |
O |
O |
|
O |
|
6 months |
O |
|
|
|
|
|
|
complication |
not specified |
liver dysfunction |
liver dysfunction |
not specified |
anemia, anorexia |
not specified |
|
additional treatment |
none |
none |
none |
surgical dissection before Huaier tr. |
chemotherapy for 4 months + radiotherapy, hormone therapy |
none |
|
prognosis |
healthy (no cancer) |
complete remission |
complete remission |
remission |
deceased (58 days) |
healthy |
Table 1: Clinical features of cancer patients and normal controls. The asterisk (*) of No.6 means that the recovery from the opportunistic RNA virus infection (asymptomatic, cured by Huaier only).
RNA extraction, miRNA library construction and mRNA library construction
The obtained peripheral blood samplesintroduced in the present studywere designated to be analyzed by RNA extraction, miRNA library construction, and mRNA Library Construction in BGI Shenzhen, followed by the construction of whole transcriptome library for sequencing on BGISEQ-500 Platform, followed by whole transcriptome mapping [25]. The detailed methodology was described precioussly, and theprotocols including bioinformatics workflow were provided and demonstrated at BGI.
In the transcriptome analysis, all samples had library construction and sequenced on the MGISEQ-2000 and BGISEQ-500 platform. Paired-end reads were aligned to UCSC hg38 using Hisat2 and Bowtie2. We calculated the expression of genes by RSEM, call DEGs by PossionDis method, predicted novel transcripts by StringTie and call SNP by GATK [28-34] obtained novel genes have been deposited to The NCBI GEO (GSE157086).
Finally, we identified DEGs (differentially expressed genes) between samples and do clustering analysis and functional annotations. With quantitative analysis of DEGs, we performed Gene Ontology (GO) classification, functional enrichment, and KEGG pathway classification [35] according to each cancer analyzed in the present study and functional enrichment with the KEGG annotation results then classify DEGs according to official classification, and were also perform pathway functional enrichment (only significant one). Furthermore, we applied the enrichment analysis of DEG in KEGG database.
Small RNA analysis
We have also performed total non-coding small RNA sequencing on the same BGISEQ-500 Platform, and the length of the small RNAs were found to be between 18 to 30 nucleotides to make unique small RNA mapped to one annotation following priority rule: miRNA>piRNA>Rfam>other sRNA. After sRNA annotation, those unknown tags will be used to predict novel sRNA based on their architectural features. Thus, novel miRNA, piRNA and siRNA were predicted [25,36-38].
The following processes were applied to the subsequent data analysis:
- • Eliminate the low-quality reads, small tags, adaptors and other contaminants to get clean reads
- • Summarize the length distribution of the clean tags, common and specific sequences between samples
- • Annotate the clean tags into different categories
- • Predict the novel miRNA by exploring the characteristic hairpin structure of miRNA precursor. E. Function annotation of known miRNAs, including Gene Ontology and Pathway
The obtained novel small nuclear non-coding RNA have been uploaded to the former deposited information; The NCBI GEO (GSE157086) [25].
Results
Summary of sequence events
The quantitative analysis of > 7.0 GB RNA sequencing of each sample provided estimated transcribed genes of >27,000; mean number of 27, 447 per sample, and the average mapping ratio with reference genome is 89.64 % (varies from 71.49% to 94.51%) as previously reported for the present study, additional 22, 400 transcripts were identified and 15,430 of them are already known genes, 4,008 of them are novel genes, and the rest 3,062 were non-coding RNAs. The obtained novel genes and small nuclear non-coding RNA obtained in the present study have been up-loaded to the genes already deposited to the NCBI GEO (GSE157086) [25].
SNP variants indicated no significant relationship to the fate of the patients
Figure 1 demonstrated the SNP variants compared between the samples obtained before and after 1month, and 3 months and the follow-up monitoring after 1year of Huaier administration (20g per day) [25,38]. We identified total 1,610,505 SNP variant types in 18 samples used in the present study, and 89,473 Single Nucleotide Polymorphism (SNP) variants per sample in mean number (ranging from 59,392 to 125,975), where as 22,688 in total among normal healthy individuals [39]. As for the SNP variations, A-G > C-T transitions were the most common mutations (593,513and 591,446 respectively, 73.5%), followed by A-C > A-T (10,7051 and 80,821, respectively, 11.7%), C-G > G-T (132,972 and 104,702 respectively, 14.8%) transfusions, which is consistent with the previous reports on oesophageal squamous carcinoma cells [24].
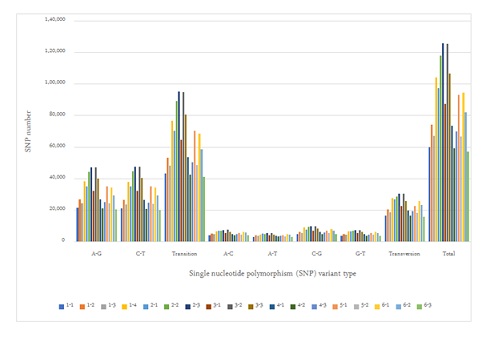 Figure 1: SNP variance after Huaier administration; classifications by SNP variation types.
Figure 1: SNP variance after Huaier administration; classifications by SNP variation types.
These ratios found no significant differences among samples, and the distribution and the numbers in each alteration was almost the same as the result reported previously [25].There were no significant differences identified by the stages of cancer. Distribution of both SNP and INDL (>2,000,000 per sample) location was identified 40 to 45 % in exons and up to 60% including the adjacent area. Thus, in contrary to the previous reports, these RNA editing events did not randomly scatter among the whole genome, or any correlation to cancer origin or stages [40,41].
Genome-scope view of detailed alterations pattern recognized b ydifferentially expressed genes DEGs), transcription factor-DEG network and KEGG pathway analysis [25]
Transcribed gene analysis enabled us to compare the levels of Differentially Expression Genes (DEG) in 18 samples used in the present study by the time course of Huaier administration (Figure 2). Comparison of up- and down-regulated transcriptomes by numbers was shown in figure 2. Each bar represents the comparison before and after 30 days and 90 days then indicated months after Huaier administration, the research period has been set to be 90 days, but all the patients continued 20g per day for one year, and gradually reduced the quantity for 3g per day use (patient No.1 and 4).
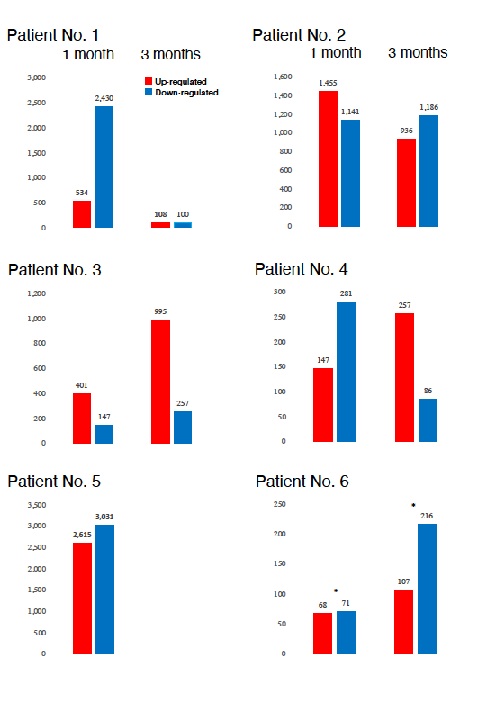 Figure 2: Comparison of the numbers of differently expressed genes (DEGs) in each patient by time course of Huaier administration. The asterisks* indicate the time of opportunistic virus infection.
Figure 2: Comparison of the numbers of differently expressed genes (DEGs) in each patient by time course of Huaier administration. The asterisks* indicate the time of opportunistic virus infection.
Huaier effects on transcriptional factors and their quantitative rearrangements in vivo were shown in figure 3. The relationship of those quantitative and qualitative DEG alterations to encoding Transcription Factors (TF) was clearly demonstrated by the TF-DEG networkchanges [25]. This functional linkage map of TF-DEG network was most convenient form to comprehend the level of genetic changes and the course of recovery. The red and green dots represent the up-regulated and down-regulated DEGs, respectively. Purple ball represents each TF as a core. The greater the core size, the more DEGs regulated by the core transcription factor. The results indicated the changes in gene expression by Huaier administration were resulted in, and also caused by altered transcription control.
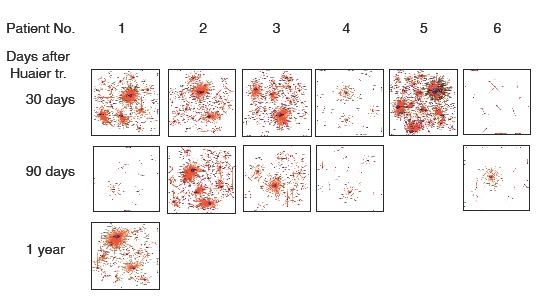 Figure 3: Genome-scope view of TF-DEG network in each patient by the time course of Huaier administration. The red and green dots represent the up-regulated and down-regulated DEGs, respectively. Purple ball represents transcription factor, the greater the node the more DEGs the transcription factor regulate.
Figure 3: Genome-scope view of TF-DEG network in each patient by the time course of Huaier administration. The red and green dots represent the up-regulated and down-regulated DEGs, respectively. Purple ball represents transcription factor, the greater the node the more DEGs the transcription factor regulate.
The volunteer patients and a normal control were free from any conventional anti-cancer chemotherapy during the research period up to present. TNM Stage IV patient (No.5) was first diagnosed as or pharynx squamous cell carcinoma, further examinations revealed additional cancer; hepato-cholangiocarcinoma and prostate cancer. After 100 days of chemotherapy (Cisplatin: CDDP, Bevacizumab and etc.) hormone therapy and radiotherapy, the patient decided to switch to have Huaier therapy. After 30 days of Huaier administration, the assessment of QOL was significantly improved, increased appetite, nutrition level, hemoglobin counts (6.0 to 10.0 g/dl).However, the physiological conditions gradually got worse, and deceased at 58 days after Huaier therapy. During those 58 days, it was no need to use morphine anti-pain treatment.
Huaier administration (20g per day), by dose and duration, could completely rescue the molecular functions to the normal level (to recover homeostasis).The required time for the recovery seemed to vary among patients, from 30 days to several months, and severe case such as patient No.5 could not recover from the immunodeficiency. However, even in this case, physiological status has much improved, judging from the estimation by the assessment of the patient QOL, and with the medical check in the clinics (such as hemoglobin titers).
Figure 4 demonstrated detailed factors and molecules with up/down regulation according to the KEGG pathway map [35]. Here we need typical pattern recognition by genome-scope view, and compare the molecule dynamics involved in pathway functions. Immunological improvements seemed to be closely linked to the regulation of NFkB (nuclear factor kappa-light-chain-enhancer of activated B cells) signaling pathways, which also closely related to improve prognosis. The panels represented Huaier effects on each patient (noted by upper left) after 30 days and 90 days after Huaier administration, with alterations of DEGs indicated by red colored box (up-regulation) and blue-colored box(down-regulation).In each panel, the specified DEG names were highlighted in grey box, especially related to oncogenes and tumor suppressor genes.
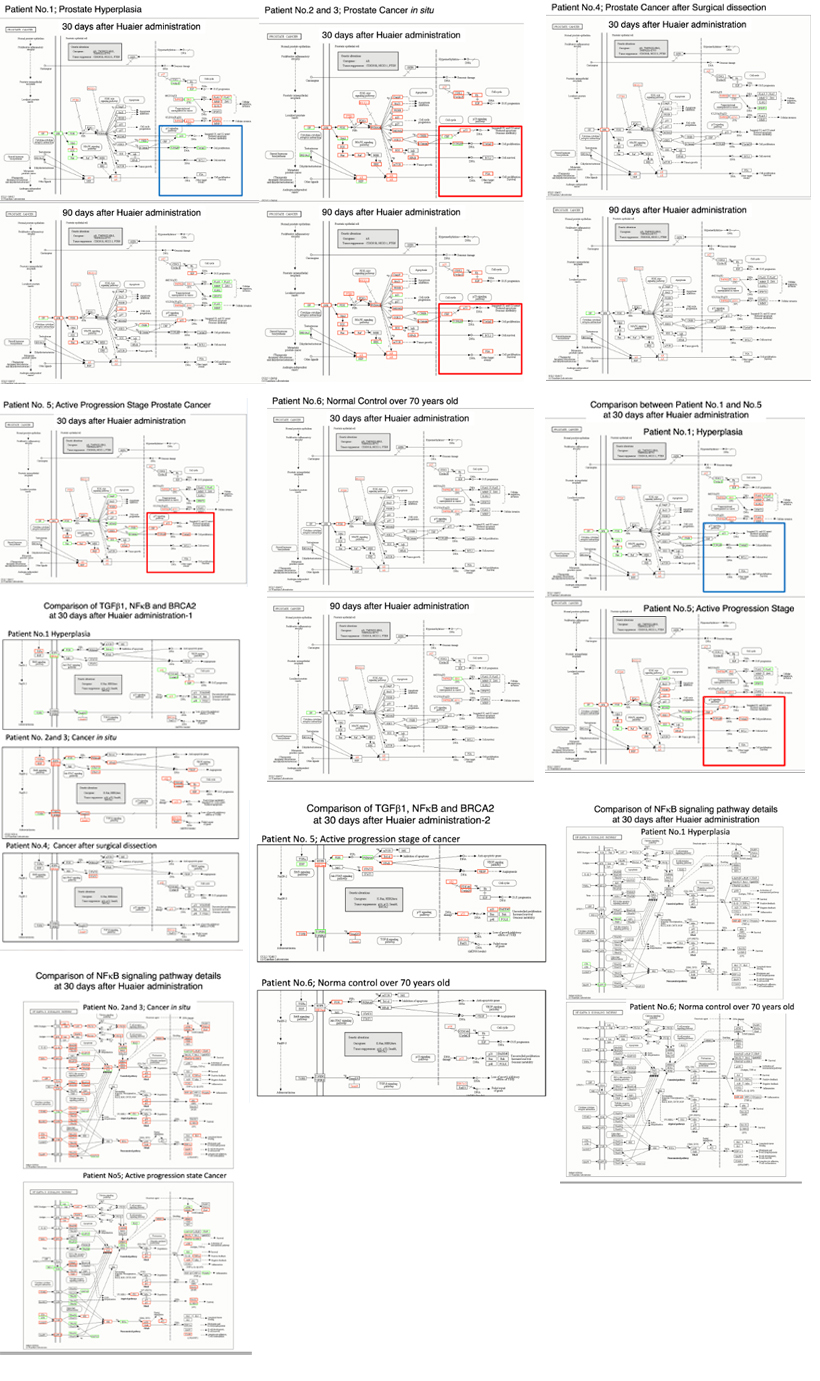
Figure 4: Genome-scope pattern recognition of Huaier effects on each biophisiological systems by KEGG biological pathways [35]. Since a broad range of enriched pathway with various up- and down-regulation of transcripts involved, KEGG pathway classifications are convenient to recognize alterations among patients, by the time course of Huaier administration. The pathways were classified so as to cover all the molecular function categories, i.e., every process involved in molecular biological function, cellular component and biological process. The pathways responsible for prostate cancer progression, stress reduction, and major immunological responses related to p53, NFkB and TGFb signaling pathway including BRCA2 function were specifically compared among patients with hyperplasia, cancer in situ (all recovered or in remission stage), and with poor prognosis. The altered oncogenes and tumor suppressors were highlighted in the box in each panel.
We searched for the possible molecular changes by the comparison between the patients recovered and deceased (patients number noted in Table 1were written on the left side of each panel).The representative changes were highlighted in grey box such as: Oncogenes; c-Myc, TERT, NRF2, c-Met, PIK3CA, k-Ra, HER2/neu, EGFR, MDM2, PDGF, PDGFR, CDK4; Tumor suppressors; Rb, p53, PTEN, p16, Smad4, BRCA2, IKN4a/ARF, AXIN, KEAP1, ARIDIA and DNA repair genes; hMLH1, hMSH2, hMSH3, hMSH6.
The up-regulation of DEGs responsible for the signal transfer in p53, NFkB and TGF-b (transforming growth factor beta) signaling pathway [35] showed strong correlations for inhibition of cancer progression, and relapse after surgical dissection, with further influences to related regulatory systems.
Not only the genes correlating with signal transduction and carcinogenesis, many other genes of physiological importance should be also affected as a link to main pathways for maintenance of homeostasis. More importantly, almost all the transcriptional misregulation was rescued chiefly by the up-regulated transcription factors, with the down-regulation of Nuclear receptor co-repressor (N-CoR) in Transcriptional Misregulation in Cancer [25]. As reported before [5], figure 3b confirmed the transcription control rescue in Hippo signaling pathway [3] and corresponding Wnt and MAPK signaling pathways.
MiRNA-mediated alterations in transcription control:Analysis of differentially expressed small RNAs (DSGs); siRNA (30 nucleotides), piRNA (24 nucleotides) and miRNA (21-22 nucleotides)
The numbers of detected small non-coding RNA for each sample were summarized in figure 5. The structure of miRNAs and the sequences of the novel small RNAs were deposited to the NCBI GEO (GSE157086), and shown and stored in precursor.tar.gz.
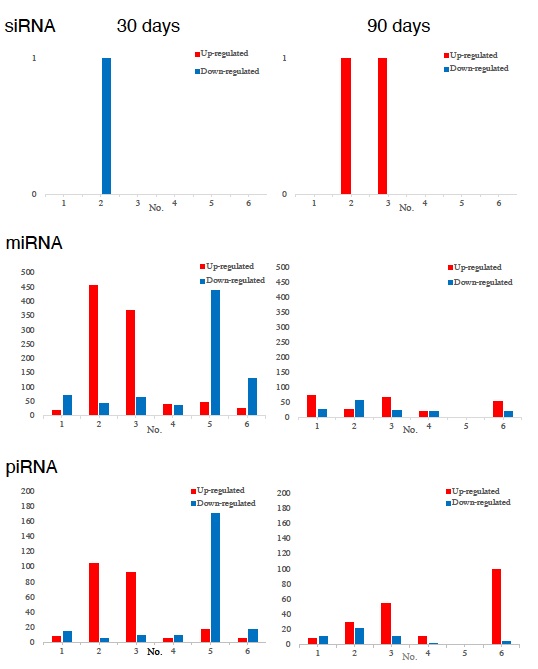 Figure 5: Comparison of the numbers of non-coding small RNAs. Upper: Statistics of siRNA, middle: miRNA, lower: piRNA.
Figure 5: Comparison of the numbers of non-coding small RNAs. Upper: Statistics of siRNA, middle: miRNA, lower: piRNA.
Compared with the changes in transcriptomes (Figure 2) and TF-DEG network (Figure 3), miRNAs seemed to contribute to the drastic alterations of down regulation of the transcriptomes, followed by up-regulation to cease the overall alterations in the end, i.e., after recovery from the disease. Figure 5 well demonstrated miRNA-mediating gene silencing effects with a comparison to the results in figure 2. For example, the significant changes in up- and down-regulated DEGs were in inverse proportion to those observed in quantitative DES changes, such as significant increase of up-regulated miRNAs in the patient No. 8 resulted in the significant decrease of both up- and down-regulated DEGs. In contrast, the significant decrease of up-regulated DESs in the patient No.6, related to human papillomavirus infection, drastically increased both up- and down-regulation of DEGs up to 3,330 and 3,505, respectively (12.2% of the total DEGs).
Thus, miRNA-mediated were directly associated with the drastic up-regulation of transcription factor families, which resulted in the modifications in DEGs.
SiRNA seemed to have less correlations with general alterations observed in the present study, and drastic functional changes were found in oesophageal carcinoma in the other patient. In the present study, no significant siRNA function was identified so far.
In the present study, haemoglobin counts in TNM stage IV multi-cancer patient improved from 6.0 to 12.0 g/dl MiRNAs also play crucial roles in the regulation of complex enzymatic cascades including the haemostatic blood coagulation system [42,43]. Large scale studies of functional miRNA targeting have recently uncovered rationale therapeutic targets in the haemostatic system [43].
Piwi-interacting RNA (piRNA) is the largest class of small non-coding RNA molecules (containing about 24 nucleotides in the present study) expressed in animal cells [44]. PiRNAs were reported distinct from microRNA (miRNA) in size (24 nucleotides as opposed to 21–22 nt), lack of sequence conservation, increased complexity. However, as shown in figure 5, the dynamic statistics of piRNA revealed similar movement as miRNAs, although the total numbers of the known and the novel piRNAs were much smaller compared with miRNAs [45]. The present study, together with other small RNA information, shed lights into the function of piRNA function related to RNA-mediated adaptive against transpose on expansions and invasions, especially in the case with the opportunistic infection [46].
Discussion
The results obtained in the present study clearly indicated anti-cancer effects of Huaier, by successful rescue of genetic functions and related genes which contributed to carcinogenesis. These genetic alterations were based on the rescue of transcription control, which contributes to maintain and restore homeostasis; individual genomic potential equally played an important role to undergo these drastic changes.
As shown in figure 1, the average number of RNA editing events such as SNP and INDL showed that Huaier administration induced significant increase in movability of the genome, ranging 2 times to 7 times higher than those detected as identified among normal population [39]. The statistics of splicing events resulted in expanded the plasticity, flexibility, and consequently enhanced the genomic capability to manage or cope up with the transcription and translation. However, any significant link was not detected for the progression and prognosis of prostate cancer [40,41].
Compared with the other malignant cancers reported previously [25], the genome-scope view in figure 4 indicates that the genetic alterations caused complete remission of prostate cancer were not so large in quantity thus alterations were much larger in quantity in healthy control, and that at the time of opportunistic virus infection. However, those minimum essential modifications were enough to cause required recovery were observed in prostate cancer. It is surprising to see genomic flexibility and capability, i.e., individual genome potential which can tolerate extensive genomic and genetic changes. The typical feature was observed in the most quantitative DEG changes observed in the patient 5 at 30 days after Huaier administration. Severe chemotherapy, hormone therapy and radiotherapy before Huaier administration seemed to diminish potential and genomic energy. The effect of conventional chemotherapy, especially about the genome-scope analysis of Cisplatin: CDDP and FOLFORINOX treatment, have already obtained. It appeared about 3 months after administration and surely limits the genomic potential (data nor shown here) then we recommended to use Huaier first and then surgical dissection (according to the stage), and followed by Huaier therapy at the best for progressive active phase of prostate cancer.
These changes in transcripts were successfully demonstrated as Transcription Factor (TF)-Differentially Expressed Genes (DEG) lineage map (Figures 3 and 4).This panel is suitable to evaluate the recovery process by the time course of Huaier administration, and it is also true to see the recovery from the influence of environmental stress typically indicated by the opportunistic virus infections observed in normal healthy controls, too. The duration period required for the recovery was dependent on the physiological status in each individual, one month for healthy control and about 5-6 months for cancer patients. This observation was identical to the clinical observation reported previously.
The comparison between the patients who successfully recovered and failed revealed the major difference existed in immunological factors, especially observed in the control in p53, NFkB, TGF-b and BRCA2 signalling pathways (Figure 4).The cancer recovery seemed to require practical inhibition and down-regulation of NFkB and TGF-b and functionally linked molecules after significant up-regulation. These sequential changes occurred by time course of Huaier administration and other signalling pathways. The sequential control of immunological reactions, both activation as well as modulation of excessive enhancement, were required for the recovery of cancer, by NFkBand TGF-b signaling pathways.
On the other hand, p53 and p53 signaling pathways seemed to have more influence for the progression of cancer from hyperplasia (Figure 4).The disruption in p53 signaling pathway in various cancers was frequently observed among the volunteer patients in our genome-scope projects. In addition, it is emphasized that genomic plasticity has been continuously observed even in normal healthy individuals. It seems that, even without any medical problems and disorders, every person should have influences from environmental stress, infective agents and ageing processes which disrupted physiological functions. Huaier revealed its efficacy even on the healthy individual, according to the extent of functional disruption. The observation provided of the potential of Huaier to rescue the silent disruptions in molecular level to recover the normal homeostasis. The recovery of cancer was easily identified to see the quantitative level of regulation in transcriptomes, with a confirmation of CT, MRI, and X-ray images of the patients (data not shown).However, there were many patients without significant gene expression changes.
Huaier administration induced as such many clusters of oncogenes and tumor suppressor genes, and that in both up- and down-regulated ways.It seems very typical and of great importance to enhance anti-cancer immunity by Huaier, which was maintained within essenatial level, not to become excesive in amount.There have not been reported yet about the possible compounds or natural herbs to show the similar efficacy in vivo. Many trials were performed, but failed from the strong toxicity even to in vitro cultured cells.In contrast, Huaier seemed to compensate for the limitation of targeting system by conventional chemotherapy, by the rescue of all the required physiological functions.Clinically, the rescued functions seemed to be responsible for the specific cell death in occult metastasis in the peripheral blood (prevent the recurrence and relapse of cancer) and for the recovery from anemia and hypoleukocytemia.
The present study also increased the information of the many novel miRNAs significantly contributed to cancer recovery, and further investigation of these molecules in vitro and in vivo are expected (Figure 5). MiRNAs have been reported to typically silence genes by repression of translation, and with broader specificity than siRNA, to functions in RNA silencing and post-transcriptional regulation of gene expression [25,42-46]. The consequent gene-silencing were also detected by miRNA-mediated post-transcription control and numerous novel molecules were identified (deposited to NCBI GEO: GSE157086).
In addition, we emphasized the significance of Huaier effects to the opportunistic infections, such as RNA virus infection as COVID-19.The severity of symptoms and the recovery period dependent on the genomic potential, too, and in fact, the level of severity to the lethal situation in case were drastically decreased by clinical trial of Huaier against COVID-19 (WHO ID:NCT04291053). Huaier also prevented reinjection after 3-6 months (data not shown).
Conclusion
The present study provided Huaier effects on the inhibition of active carcinogenesis and prevention of relapse in prostate cancer, with the molecular basis of miRNA-mediated transcriptional control. The recovery from cancer was identical to the recovery of homeostasis, based on the modulation and rescue of each process in transcription and translation. However, no specific RNA events, SNP variations were detected as a significantly link to cancer remission, or prevention of relapse in prostate cancer. Surgical dissection or other therapies can be avoided by Huaier conventional therapy, together with cancer-specific monitoring system by blood test and practical imaging. The individual genomic potential to undergo those changes and modulations is the key for cancer recovery.
Acknowledgement
The authors wish to thank cancer patient volunteers and many healthy volunteers kindly collaborated with the present study. The present study was grant-in-aid from QiDong Gaitianli Medicines Co., Ltd. And Japan Kampo New Medicine, Co., Ltd.
Author contributions
T.T., M.T., designed the study from the clinical observation of the cancer patients with Huaier treatment (as a complementally therapy), and managed the sampling and clinical assessment of the patient volunteers, statistically analyzed the data, and drafted the manuscript. F.T., H.L., managed total RNA and small nuclear RNA sequencing and conducted systematic analysis of the data. Z.L., D.W., contributed to the provision of Huaier granules and clinical evaluation of the data, especially focused on Immunological evaluation.
Conflict of Interest
The authors have no competing interest to declare.
References
- World Cancer Report 2014 (2014) International Agency for Research on Cancer. World Health Organization, Geneva, Switzerland.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin 68: 394–424.
- Alberts AR, Schoots IG, Roobol MJ (2015) Prostate-specific antigen-based prostate cancer screening: Past and future. Int J Urol22: 524-532.
- Wallis CJ, Glaser A, Hu JC, Huland H, Lawrentschuk N, et al. (2018) Survival and Complications Following Surgery and Radiation for Localized Prostate Cancer: An International Collaborative Review . Eur Urol73: 11-20.
- Tanaka T, Kawamura Y, Usui Y, Terachi T, Kimura F, et al.(2010) Bradeion Project: Monitoring and Targeting of Cancer: Molecular Marker Diagnosis of Cancer by Fluorescence Correlation Spectroscopy (FCS).The Open Conf Proc J 1: 129-137.
- Tanaka M, Tanaka T, Kijima H, Itoh J, Matsuda T, et al. (2003) Rapid and quantitative detection of a mammalian septin Bradeion as a practical diagnostic method of colorectal and urologic cancers. Med Sci Monit 9: 61-68.
- Garcia W, de Araújo APU, Lara F, Foguel D, Tanaka M, et al. (2007) An intermediate structure in the thermal unfolding of the GTPase domain of human septin 4 (SEPT4/Bradeion-beta) forms amyloid-like filaments in vitro. Biochemistry 46: 11101-11109.
- Garcia W, de Araújo APU, de Oliveira Neto MO, Ballestero MRM, Polikarpov I, et al. (2006) Dissection of a human septin: definition and characterization of distinct domains within human SEPT4. Biochemistry45: 13918-13931.
- Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, et al. (2018) MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med 378: 1767-77.
- Wang X, Bao J, Ping X, Hu C, Hou J, et al. (2018) The diagnostic value of PI-RADS V1 and V2 using multiparametric MRI in transition zone prostate clinical cancer. Oncol Lett16: 3201-3206.
- Song X, Li Y, Zhang H, Yang, Q (2015) The anticancer effect of Huaier (Review). Oncol Rep 34: 12-21.
- Wang X, Wang N, Cheung F, Lao L, Li C, et al. (2015) Chinese medicines for prevention and treatment of human hepatocellular carcinoma: current progress on pharmacological actions and mechanisms. J Integr Med 13: 142-164.
- Tsukagoshi S, Hashimoto Y, Fujii G, Kobayashi H, Nomoto, K, et al. (1984) Krestin (PSK). Cancer Treat Rev11: 131-155.
- Tanaka T, Suzuki T, Nakamura J, Kawamura Y, Sadahiro S, et al. (201) Huaier Regulates Cell Fate by the Rescue of Disrupted Transcription Control in the Hippo Signaling Pathway. Arch Clin Biomed Res1: 179-199.
- Mo JS, Park JW, Guan KL (2014)The Hippo signaling pathway in stem cell biology and cancer.EMBO Rep 25: 642-656.
- Zhang H, Lang TY, Zou DL, Zhou L, Lou M, et al. (2019) miR-520b Promotes BreastCancerStemness Through Hippo/YAP Signaling Pathway. Onco Targets Ther 12: 11691-11700.
- Wu L, Yang X (2018) Targeting the HippoPathway for BreastCancerCancers (Basel) 10:422-437.
- Wei C, Wang Y, Li X (2018) The role of Hipposignal pathway in breastcancerOnco Targets Ther 11: 2185-2193.
- Feng X, Zhang M, Wang B, Zhou C, Mu Y, et al. (2019) CRABP2 regulates invasion and metastasis of breastcancerthrough hippo pathway dependent on ER status.J Exp Clin Cancer Res 38: 361-368.
- Shen J, Cao B, Wang Y, Ma C, Zeng Z, et al. (2018) Hippocomponent YAP promotes focal adhesion and tumour aggressiveness via transcriptionally activating THBS1/FAK signalling in breastcancer.J Exp Clin Cancer Res 37: 175-191.
- Qiao K, Ning S, Wan L, Wu H, Wang Q, et al. (2019) LINC00673 is activated by YY1 and promotes the proliferation of breastcancercells via the miR-515-5p/MARK4Hippo signaling pathway. J Exp Clin Cancer Res 38: 418-432.
- Wang S, Su X, Xu M, Xiao X, Li X, et al. (2019) Exosomes secreted by mesenchymal stromal/stem cell-derived adipocytes promote breastcancercell growth via activation of Hippo signaling pathway. Stem Cell Res Ther 10: 117-128.
- Jiang Y, Sun A, ZhaoY, Ying W, Sun H, et al. (2019) Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 567: 257-261.
- Song Y, Li L, Ou Y, Gao Z, Li E, et al. (2014) Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509: 91-95.
- Tanaka M, Tanaka T, Teng F, Lin H,Li N, et al. (2020) Huaier Induces Cancer Recovery by Rescuing Impaired Function of Transcription Control Based On the Individual Genomic Potential. Arch Clin Biomed Res 4: 817-855.
- Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, et al. (1997). The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med 336: 1401-1408.
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, t al. (2013). Punctuated evolution of prostate cancer genomes. Cell 153: 666-677.
- Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. (2009) The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 38: 1767-1771.
- Kim D, Langmead B, Salzberg SL. (2015) HISAT: a fast spliced aligner with low memory requirements Daehwan HHS Public Access. Nat Methods 12: 357-360.
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, et al. (2011) Integrative Genome Viewer. Nat Biotechnol 29: 24-26.
- Pertea M, Pertea GM, Antonescu CM, Chang T, Mendell JT, et al. (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290-295.
- Trapnell C (2013) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562-578.
- Kong L, Zhang Y, Ye Z, Liu X, Zhao S, et al. (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: 345-349.
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491-498.
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: 480-484.
- Griffiths-Jones S (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: 140-144.
- Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, et al. (2015) Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res 43: 130-137.
- Shen S, Park JW, Lu Z, Lin L, Henry MD, et al. (2014) rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Natl Acad Sci USA111: 5593-5601.
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, et al.(2012) Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 30: 253-260.
- Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, et al. (2008) Multiple newly identified loci associated with prostate cancer susceptibility. Nature Genet 40: 316-321.
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, et al. (2008) Multiple loci identified in a genome-wide association study of prostate cancer. Nature Genet 40: 310-315.
- Teruel-Montoya R, Rosendaal FR, Martínez C (2015) MicroRNAs in hemostasis. J Thromb Haemos 13: 170-181.
- Nourse J, Braun J, Lackner K, Hüttelmaier S, Danckwardt S (2018) Large-scale identification of functional microRNA targeting reveals cooperative regulation of the hemostatic system. J Thromb Haemos 16: 2233-2245.
- Seto AG, Kingston RE, Lau, NC (2007) The Coming of Age for Piwi Proteins. Mol Cell 26: 603-609.
- Bartel DP (2009) MicroRNAs: Target Recognition and Regulatory Functions. Cell 136: 215-233.
- Ma L, Cao J, Liu L, Du Q, Li Z, et al. (2019) Lncbook: A curated knowledgebase of human long non-coding rnas. Nucleic Acids Res 47: 128-134.
Citation: Tanaka M, Tanaka T, Teng F, Lin H, Li N, et al. (2021) Anti-Cancer Effects of Huaier on Prostate Cancer; miRNA-Mediated Transcription Control Induced Both Inhibition of Active Progression and Prevention of Relapse. J Altern Complement Integr Med 6: 146.
Copyright: © 2021 Manami Tanaka, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

