
Arthropod-Borne Disease May Lead to a More Resilient Type I Interferon Response to Coronavirus by Reducing Genetic Defects of Innate Immunity by Natural Selection
*Corresponding Author(s):
Martin BalzanConsultant Respiratory Physician, Mater Dei Hospital, Dun Karm Street, MSD 2090, Malta
Email:Martin.v.balzan@gmail.com
Abstract
This article builds on a previous proposal of a hypothesis whereby repeated arthropod or sand-fly vector infection of humans by novel viruses of zoonotic origins carrying bat or mammalian RNA/DNA, such as phleboviruses may have resulted in the development of an effective evolutionary immune response to most novel zoonotic viruses such as SARS-CoV-2 through survival of the fittest possibly over many generations. Phleboviruses and Flaviviruses such as Dengue Fever virus (DENV), can infect many mammalian species, including bats and animal husbandry just like coronaviruses. The resulting mutations when crossing species can expose large populations of humans to novel viruses that find the reactive immunity completely unprepared. Survival over thousands of years of human evolution was likely to be dependent on strong and resilient innate immunity, in the form of the Interferon I system.
Phleboviruses and Flaviviruses just like Sars-Covid-1 and 2, have multiple biochemical pathways to suppress the IRF3/7 pathway on different levels, which lead to inefficient production of type 1 interferons and an impaired antiviral response. Recently published research has shown that 3.5% of patients with life-threatening COVID-19 pneumonia had known AR IRF7 and IFNAR1 deficiencies or AD TLR3, TICAM1, TBK1, and IRF3 deficiencies or new (AD UNC93B1, IRF7, IFNAR1, and IFNAR2 deficiencies) genetic defects in the TLR3- and IRF7-dependent induction and amplification of type I IFNs.
In this paper, it is being proposed that populations of the geographical areas of the respective arthropod-borne disease such as phlebovirus or flavivirus infections may have contributed to select within the populations' survival of individuals free from genetic defects of these pathways.
Genetic epidemiological studies determining the prevalence of deficiencies in Type I interferon genes in the areas where phleboviruses/flaviviruses are present and a comparison with areas of high case fatality such as Lombardy, the United Kingdom would be necessary to support such a hypothesis.
Keywords
Corona virus; Immunity; IgM; Antibody
Introduction
Low prevalence of Mortality and case fatality rates to Sars-Covid-2 were noted in many areas of southern Europe particularly in areas where phlebotomous sandflies and phlebovirus infections are prevalent [1]. Phleboviruses carry bat, artiodactyl, and rodent RNA showing an ability to mutate and evolve across mammalian species, in a way similar to human coronaviruses [2]. A possible hypothesis was proposed by this author that by a process of natural selection after exposure of such novel viruses over many generations it is possible that the innate immune system could be more protective against novel viruses such as Sars-Covid-2 [1].
A Brazilian study (still in pre-print) reported that many regions showed significant negative correlations between Sars-Covid-2 incidence, infection growth rate, and mortality to the percentage of people with antibody (IgM) levels for dengue fever [3]. Another study by Silvestre et al reported lower mortality for Sars-Covid-2 in a group of patients with a previous history of dengue when compared with those who had not, and suggested that Dengue fever might induce immunological protection against severe acute respiratory syndrome in patients with Sars-Covid-2 infection [4].
Japan and South Korea reported low incidence and low case fatality rates similar to southern European countries [5]. The authors suggest a number of explanations such as organisation of public health, and discipline within the citizens, and the low number of ACE 2 receptors among the Asian population. One other possibility which the author of this paper proposes is that the prevalence of the Japanese B encephalitis virus may be responsible. JBE is a flavivirus just like dengue, caries RNA from animal species such as bats, birds, pigs, and horses [2,6] and like phlebovirus, and Coronavirus can mutate to novel viruses across species. It is possible that by natural selection over many generations the effectiveness of human innate immunity is rendered more competent and responsive to novel zoonotic viral infections.
Zhang et al reported that at least 3.5% of patients with life-threatening Sars-Covid-2 pneumonia had known or new genetic deficiencies of the innate immune system, more specifically the type one interferon system mostly related to the double-stranded RNA sensor TLR3- and IRF7-dependent induction and amplification of type I IFNs [7]. It appears that a robust IFN I response is necessary for the body to overcome Sars-Covid-2 infection [8].
Type-I IFNs play a major role in innate immunity during viral infections. The body must rely on innate interferon-mediated defences to limit pathogen replication and allow sufficient time for the development of critical humoral protection [9]. Cells stimulated through IFNAR induce various IFN-stimulated genes (ISGs) to repress viral replication. IFNB gene promoter activation is induced via the coordinated binding of the homodimer of interferon regulatory factor 3 (IRF3) (or the heterodimer of IRF3 and IRF7), nuclear translocation of NFkB, and ATF2/cJun (AP1) to the positive regulatory cis-elements for IFNB gene [9,10].
The aim of this paper is to further update the hypothesis where arthropod Borne infections, particularly by viruses like phlebovirus and dengue which have a capacity to mutate across mammalian species, can by natural selection promote stronger innate immunity against another novel virus such as coronavirus, in particular, the IFN type I response. The possible biological mechanisms on innate immunity will be explored.
The Hypothesis
Arthropod-Borne Viruses such as phleboviruses, and flavivirus e.g. dengue fever possess the ability to cross mammalian species causing disease. This includes humans, wild animals such as bats, and animal husbandry or pets associated with humans [2]. Just like Coronavirus, this ability to cross species leads to novel infections where reactive immunity is unlikely to be present in the host, and resolution of infection and survival must rely exclusively on the innate immunity in particularly Type I Interferon production and regulation.
Phleboviruses and Flaviviruses also possess many mechanisms to block the innate naturally IFN I system [11]. Sars-Covid-2 has multiple mechanisms of blocking the natural innate interferon immunity and host survival appears to be heavily dependent on the function of the TLR7 IRF 7 pathways [7]. In areas where these arthropod-borne diseases are prevalent, because of a similar ability to produce non-structural proteins which hamper the IRF 3 pathway, genetic defects in interferon reaction particularly in the TLR7-IRF7 pathway may be wiped out from the populations over many generations by natural selection, resulting in better outcomes in a modern aeroplane-spread coronavirus pandemic probably because of a more resilient IRF 7 interferon 1 pathway.
As arthropod Borne viruses are naturally airborne they can spread disease over wide geographical areas. On the contrary prior to the age of modern, train or aeroplane transport novel coronaviruses which spread via airways or fomites were more likely to lead to localized epidemics e.g. MERS, Sars-covid-1. This has changed with modern air travel where coronaviruses can cause a pandemic particularly if they can spread effectively via the airborne route.
Of course, age and other diseases such as diabetes, or cardiovascular compromise, or immunosuppressant medication would be the main determinants of the clinical outcome of Sars-Covid-2 infections, but with a stronger baseline level of innate immunity, the outcome would also make these categories of patients more resilient to Sars-Covid-2 infection [12].
In summary, arthropod-borne viruses which have the two characteristics of mutating into novel zoonotic viruses and the ability to suppress innate immunity may make patients more resilient to Sars-Covid-2 via a process of natural selection over many generations, in particular by wiping out genetic defects of Type I interferon innate immunity.
Sars-Covid-2 Immunopathology; TLR7/IRF7 The Only Lifeline
The ability of Sars-Covid-2 to suppress the innate immune is response has been clearly illustrated by Kasuga et al [11]. as shown in Table 1 and Figure 1. A short summary of the main points is presented here. Sars-covid-2 has a number of evading strategies to evade host antiviral immunity. Once Sars-covid-2 enters a cell, the genomic RNA of coronavirus is uncoated from the nucleocapsid (N) protein, leading to the translation of two open reading frames (ORFs), ORF1a and ORF1b. This polyprotein is cleaved by NSP3 and NSP5 genes to form 16 non-structural proteins (ns1-16). NSP 3, 4, and 6 produce a double membrane vesicle to envelope viral RNA to make detection by cellular receptors difficult. ‘Pathogen-associated molecular patterns’ Receptors in the cytoplasm (PAMPs) and ‘danger-associated molecular patterns’ (DAMPs) normally are present in the cytoplasm to detect viral invasion. Toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are two major receptors responsible for sensing RNA virus infection and triggering antiviral IFN activity.
|
Coronavirus Protien |
Immune Evasion Strategy |
|
Nsp1 |
Cleaves host mRNA, Inhibits protien translation, Supresses function of STAT1 and c-jun |
|
Nsp3 |
Inhibits type I IFN prodcution, Supresses ubiquitation and ISGylation, DMV formation |
|
Nsp4 |
Sequesters viral RNA via DMV formation |
|
Nsp5 |
Processes antiviral protiens |
|
Nsp6 |
Sequesters viral RNA via DMV formation |
|
Nsp8 |
Inhibits MDA5 activation |
|
Nsp13 |
Remove 5' ppp of viral RNA |
|
Nsp14 |
RNA cap modification |
|
Nsp15 |
Remove PUN RNA of viral RNA |
|
Nsp16 |
RNA cap modification |
|
ORF 3a |
Antagonizes IFN signaling, promotes apoptosis and inflammasome |
|
ORF 3b |
Antagonizes IFN signaling |
|
p4a (orf4a) |
Antagonizes IFN signaling, antagonizes PKR function |
|
ORF 6 |
Inhibits nuclear transportation of antiviral protiens |
|
ORF 7a |
Inhibts host protein translation, active proinflammatory pathways |
|
ORF 9b |
Processes antiviral protiens, antagonizes IFN signaling pathway |
|
N |
Inhibits TRAF3-TANK-TBK1/IKK? complex formation |
|
M |
Inhibits viral sensing function of RIG-I and MDA5 |
Table 1: Immune evasion strategies by corona proteins. Reprinted from Kasuga et al. [11].
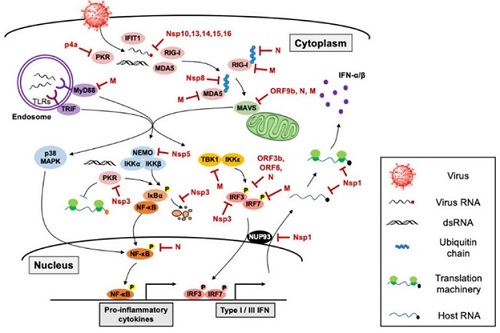 Figure 1: The host innate immune sensing pathway targeted by coronavirus. Karuga et al. [11].
Figure 1: The host innate immune sensing pathway targeted by coronavirus. Karuga et al. [11].
Studies have also shown that TLR7 plays a critical role in the sensing of coronavirus infection in certain types of immune cells. A recent clinical study discovered that several natural mutations in the TLR7 gene that cause ‘loss-of-function are associated with the severity and mortality of young Sars-COVID-2 patients, demonstrating that TLR7 sensing of SARS-CoV-2 is critical for the control of Sars-Covid-2 pathogenesis [13]. Although SARS-CoV-2 infection can suppress the type I and III IFN signalling pathways, pro-inflammatory responses are still robustly induced during SARS-CoV-2 infection [14] mainly through the TLR7 and IRF7 pathway. It is likely that SARS-CoV-2 is also sensed by these RLRs. RNA sensing TLRs, TLR3, TLR7, and TLR8, are located in the endosomal membrane and detect double-stranded RNA (dsRNA; TLR3) and single-stranded RNA [14].
Nsp1 of coronaviruses has been reported to impede the host innate immune system by targeting multiple biological pathways, such as Nsp1 inhibiting the translation of host antiviral proteins by targeting the 40S ribosomal subunit, either blocking the assembly of the translation machinery or inducing the cleavage of 5′-capped host mRNAs [15,16] and suppresses protein translation by inducing 5′-capped host mRNA cleavage, blocks mRNA nuclear export by directly inhibiting nuclear pore complex protein 93 (Nup93), suppressing protein synthesis [17,18].
SARS-Cov-2 Nsp3 can bind to IRF3 and prevent the phosphorylation, dimerization, and nuclear translocation of IRF3, resulting in the suppression of the IFN 3 signalling pathway [19]. The whole impressive array of known interference with the innate immune system is illustrated in table 1 [11] and is beyond the scope of this paper. However, the main point is that the outcome of this battle between the virus and cells of the human body, in particular with inhibition of the IRF 3 pathway appears to be mainly determined by the efficacy of the TLR7-IRF7 system’s ability to stimulate the interferon I system sufficiently, to overcome the virus or at least delay the process until re-active immunity kicks in. Kasuga et al summarize it “Failure of timely and appropriate innate immune activation may lead to robust viral propagation and is directly associated with disease severity and mortality’’ [11].
Phlebovirus Non-Structural Proteins. Blockage of the IRF3 Pathway
Wuerth et al. [20,21] have demonstrated that the Sicilian Sand-fly Virus (SFSV) suppresses the induction of the antiviral type I interferon (IFN) system in an NS-dependent manner. SFSV NSs interfere with the TBK1-interferon regulatory factor 3 (IRF3) branch of the RIG-I signalling pathway but not with NF-κB activation. In contrast to IRF3, neither the IFN master regulator IRF7 nor any of the related transcription factors IRF2, IRF5, and IRF9 were bound by SFSV NSs. The authors also state that inhibition of promoter binding by IFN transcription factor 3 (IRF3), is a molecular strategy that is unique among phleboviruses and, among human pathogenic RNA viruses in general [20].
Although the generation of a full IFN response has been attributed to the IFN-inducible IRF7 [22], the IRF3 pathway is indispensable for the induction of the first wave of IFN-α expression from virus-infected cells and [23].
Sand fly-borne Toscana virus TOSV NSs have been shown to target IRF-3, preventing its dimerization and, hence, the activation of IFN transcription [24]. Furthermore, a genetically modified (TOSV), where the non-structural protein was not expressed TOSV was unable to (i) suppress interferon (IFN)-b messenger RNA induction; and (ii) grow efficiently in cells producing IFN-b [25].
Rift Valley fever virus (RVFV) is another phlebovirus that caused an outbreak in Egypt in1977-78, where there were an estimated of 20,000 to 200,000 human cases and roughly 600 confirmed deaths. RVFV NSs protein is a type-I IFN antagonist, and thus supports efficient viral replication in type-I IFN-competent cells or immune-competent hosts [26].
Dengue Fever
A review by Pang et al showed 12.2% (101 out of 827) of bats with dengue virus RNA positive in urban/rural settings as compared to 5.2% (19 out of 362) among bats captured in a forest setting (p-value <0.05). Moreover, there were 1,335 dogs, 1,310 bovids (e.g. sheep, cattle, goat), 930 horses, 859 birds (including hens, ducks & geese), 563 pigs and 187 rodents reported with dengue tested positive [27].
DENV can inhibit both the production and the signalling of IFN-I. The viral proteins, NS2A and NS3, inhibit IFN-I production by degrading cellular signalling molecules. In addition, the viral proteins, NS2A, NS4A, NS4B, and NS5, can inhibit IFN-I signalling by blocking the phosphorylation of the STAT1 and STAT2 molecules. Finally, NS5 mediates the degradation of STAT2 using the proteasome machinery [28].
Dengue virus Non-structural proteins have also been shown to inhibit RIG-I/TBK1-directed IFN responses and restrict IRF3 and IFN responses and thereby facilitate DENV replication and virulence [29] DENV serotype 4 (DENV4) non-structural (NS) proteins NS2A and NS4B inhibited RIG-I-, MDA5-, MAVS-, and TBK1/IKKε-directed IFN-β transcription and dose dependently inhibited the phosphorylation ofTBK1 and IRF3 [30].
DENV actually produces an intracellular membrane that conceals dsRNA which diminishes the effect of the host defence mechanism, a strategy that differs from active suppression of IFN activity, but which is also utilized by Sars-Covid-2 [31].
Japanese B Encephalitis
JEV can infect humans and a variety of vertebrate animals including pigs, horses, birds, sheep, dogs, and birds. However, only pigs and water birds are considered reservoirs of the virus [32]. JEV and/or serum antibodies against JEV have been detected in bats in Japan and China, however, the role of bats in the JEV life cycle is unknown [32-34].
JEV NS5 inhibits IFN?I signalling inhibition through a Tyk2 phosphorylation blockade. This induces the cytoplasmic retention of STAT1/2 and prevents IRSE driven transcription. JEV NS5 may act through cellular tyrosine phosphatases to exert antagonistic effects [28,35,36].
Other Common Mechanisms amongst Flavivirus
Another mechanism of active interference with the type I IFN system is competitive binding. DENV and WNV NS3 proteins compete with RIG-I for 14-3-3ε binding, a chaperone responsible for trafficking RNA-bound RIG-I to the mitochondrial membrane [37].
Three of the most common disease-causing flavivirus-Yellow Fever, Zika, and Dengue inhibit human signal transducer and activator of transcription 2 (hSTAT2)-dependent IFN a central regulator of the type I IFN response [37].
Resolution of the Main Problem of the Original Hypothesis-No Coronavirus in Arthropods
The main problem with the original hypothesis was that coronaviruses have never been isolated from arthropods, and no arthropod RNA was ever discovered within the coronavirus genome.
In a pre-industrial scenario when, train, car, or air travel were not yet developed when coronaviruses crossed species and mutated to a novel virus unless they had been highly transmissible via the upper airways such as Sars-Covid-2, they would have probably led to localised epidemics such as Sars-Covid-1 and MERS resulting in reactive immunity or natural selection by death in a localised population. As a consequence, a modern coronavirus pandemic is less likely to be preceded by global resilience specific to coronaviruses.
On the other hand, a novel zoonotic arthropod-borne virus e.g. Phlebovirus, or Flavivirus would have caused an epidemic, eventually becoming endemic on a much wider geographical area, usually limited only by the breeding areas of the arthropod and reservoir host as determined by climatic or environmental conditions. As a result of arthropod-borne viruses, the innate immune system of a much wider population is challenged by a novel zoonotic viral illness when compared to the coronavirus. This may have resulted in more widespread future protection by the mechanism of “survival of the fittest” leading to a resilient innate immune response with fewer genetic defects.
Other Possible Problems with the Hypothesis
As stated in the original paper, the incidence of Sars-Covid-2 infection could reflect the effectiveness of public health measures and the capacity of the health care system to cope with the number of sick people during the epidemic as well as for the deployment of testing [1]. Analysis of geographic data provided for example by ECDC is not age-stratified and might be subject to errors of over or underrepresentation of specific age groups, or vulnerable population subgroups. However, the original hypothesis refers mainly to data on mortality and case fatality which tends to be more reliable and consistent in time rather than data on the total incidence of cases.
How to Test the Hypothesis
The prevalence rates of known genetic deficiencies in Type I interferon genes in the general population in the areas where phleboviruses/flaviviruses are endemic could be compared with areas where such arthropod-borne viruses are absent, and Sars-Covid-2 mortality and morbidity was high, such as Lombardy, the United Kingdom or the Netherlands. While this would prove a possible association, further studies of the RNA sequences of the viral nonstructural proteins and analysis of the chemical composition of the non-structural proteins themselves, to determine their evolutionary origin could add more weight to the possible association between arthropod-borne viruses and Sars-Covid-2 or other coronaviruses with pandemic potential.
Conclusion
Further to the previous hypothesis it is re-iterated that just like coronaviruses, arthropod-borne viruses such as phlebovirus and flavivirus can cross species such as bats and other artiodactyls and can give rise to novel zoonotic viruses. It is possible that in certain geographic areas arthropod-borne infections over many generations lead to an innate immune response that is resilient to emergent/novel zoonotic viral infections such as Sars-Covid-2. Sars-covid-2 is highly efficient in suppressing the host innate Type I interferon IRF3 response by means of Non-structural proteins. Viruses such as phlebovirus of Flavivirus which have similar, but biochemically different modes of IRF 3 inactivation could have prepared that population by leading to mortality of those individuals who are not resilient to such an assault in particular those which have genetic defects in the TLR7-IRF7 components. This process could have occurred for thousands of years over many generations. This would explain lower mortality and case fatality rates where phleboviruses and Flavivirus are endemic. However epidemiological studies on the prevalence of genetic defects of Interferon one production in the general population of these areas are needed to prove this hypothesis.
References
- Balzan M (2020) Low Incidence and Mortality from SARS-CoV-2 in Southern Europe. Proposal of a hypothesis for Arthropod-borne Herd immunity. Medical Hypotheses 143: 110121.
- Babayan SA, Orton RJ, Streicker DG (2018) Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science 362: 577-580.
- Nicolelis MAL, Raimundo RLG, Peixoto PS, de Andreazzi CS (2020) How super-spreader cities, highways, hospital bed availability, and dengue fever influenced the COVID-19 epidemic in Brazil. medRxiv.
- Silvestre OM, Costa LR, Lopes BVR, Barbosa MR, Botelho KKP, et al. (2020) Previous Dengue Infection and Mortality in Coronavirus Disease 2019 (COVID-19). Clinical Infectious Diseases.
- Iwasaki A, Grubaugh ND (2020) Why does Japan have so few cases of COVID-19? EMBO Molecular Medicine 12: e12481.
- Wang JL, Pan XL, Zhang HL, Fu SH, Wang HY, et al. (2009) Japanese encephalitis viruses from bats in Yunnan, China. Emerging Infectious Diseases 15: 939-942.
- Zhang Q, Bastard P, Liu Z, Pen JL, Velez MM, et al. (2020) Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370: eabd4570.
- Zhang Q, Bastard P, Bolze A, Jouanguy E, Zhang SY, et al. (2020) Life-Threatening COVID-19: Defective Interferons Unleash Excessive Inflammation. Med 1: 14-20.
- Stertz S, Hale BG (2021) Interferon system deficiencies exacerbating severe pandemic virus infections. Trends in Microbiology.
- Honda K, Taniguchi T (2006) IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nature Reviews Immunology 6: 644-658.
- Kasuga Y, Zhu B, Jang KJ, Yoo JS (2021) Innate immune sensing of coronavirus and viral evasion strategies. Experimental & Molecular Medicine 53: 723-736.
- Quarleri J, Delpino MV (2021) Type I and III IFN-mediated antiviral actions counteracted by SARS-CoV-2 proteins and host inherited factors. Cytokine & Growth Factor Reviews 58: 55-65.
- van der Made CI, Simens A, Schuurs-Hoeijmakers J, Heuvel G, Mantere T, et al. (2020) Presence of Genetic Variants Among Young Men with Severe COVID-19. JAMA 324: 663-673.
- Park A, Iwasaki A (2020) Type I and Type III Interferons-Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host & Microbe 27: 870-878.
- Lokugamage Kumari G, Narayanan K, Huang C, Makino S (2012) Severe Acute Respiratory Syndrome Coronavirus Protein nsp1 Is a Novel Eukaryotic Translation Inhibitor That Represses Multiple Steps of Translation Initiation. Journal of Virology 86: 13598-13608.
- Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, et al. (2020) Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369: 1249-1255.
- Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, et al. (2011) SARS Coronavirus nsp1 Protein Induces Template-Dependent Endonucleolytic Cleavage of mRNAs: Viral mRNAs Are Resistant to nsp1-Induced RNA Cleavage. PLoS Pathogens 7: e1002433.
- Gomez GN, Abrar F, Dodhia MP, Gonzalez FG, Nag A, et al. (2019) SARS coronavirus protein nsp1 disrupts localization of Nup93 from the nuclear pore complex. Biochemistry and Cell Biology 97: 758-766.
- Devaraj SG, Wang N, Chen Z, Tseng M, Barretto N, et al. (2007) Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. The Journal of Biological Chemistry 282: 32208-32221.
- Wuerth JD, Habjan M, Wulle J, Furga GS, Pichlmair A, et al. (2018) NSs Protein of Sandfly Fever Sicilian Phlebovirus Counteracts Interferon (IFN) Induction by Masking the DNA-Binding Domain of IFN Regulatory Factor 3. Journal of Virology 92: e01202-18.
- Wuerth JD, Weber F (2016) Phleboviruses and the Type I Interferon Response. Viruses 8: 174.
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, et al. (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434: 772-777.
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, et al. (2000) Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13: 539-548.
- Gori-Savellini G, Valentini M, Cusi MG (2013) Toscana Virus NSs Protein Inhibits the Induction of Type I Interferon by Interacting with RIG-I. Journal of Virology 87: 6660-6667.
- Woelfl F, Leger P, Oreshkova N, Pahmeier F, Windhaber S, et al. (2020) Novel Toscana Virus Reverse Genetics System Establishes NSs as an Antagonist of Type I Interferon Responses. Viruses 12: 400.
- Ly HJ, Ikegami T (2016) Rift Valley fever virus NSs protein functions and the similarity to other bunyavirus NSs proteins. Virology Journal 13: 118.
- Pang VJ, Gwee S (2020) Dengue in animals: A systematic review. International Journal of Infectious Diseases 101: 258-259.
- Ramirez JAC, Urcuqui-Inchima S (2015) Dengue Virus Control of Type I IFN Responses: A History of Manipulation and Control. Journal of Interferon & Cytokine Research 35: 421-430.
- Chen HW, King K, Tu J, Sanchez M, Luster AD, et al. (1950) The roles of IRF-3 and IRF-7 in innate antiviral immunity against dengue virus. The Journal of Immunology 191: 4194-4201.
- Dalrymple NA, Cimica V, Mackow ER (2015) Dengue Virus NS Proteins Inhibit RIG-I/MAVS Signaling by Blocking TBK1/IRF3 Phosphorylation: Dengue Virus Serotype 1 NS4A Is a Unique Interferon-Regulating Virulence Determinant. mBio 6: e00553-15.
- Uchida L, Espada-Murao LA, Takamatsu Y, Okamoto K, Hayasaka D, et al. (2014) The dengue virus conceals double-stranded RNA in the intracellular membrane to escape from an interferon response. Scientific Reports (Nature Publisher Group) 4: 7395.
- Liu S, Li X, Chen Z, Chen Y, Zhang Q, et al. (2013) Comparison of genomic and amino acid sequences of eight Japanese encephalitis virus isolates from bats. Archives of virology 158: 2543-2552.
- Cui J, Counor D, Shen D, Sun G, He H, et al. (2008) Detection of Japanese Encephalitis Virus Antibodies in Bats in Southern China. The American Journal of Tropical Medicine and Hygiene Am J Trop Med Hyg 78: 1007-1011.
- Miura T, Kitaoka M (1977) Viruses isolated from bats in Japan. Archives of Virology 53: 281-286.
- Cumberworth SL, Clark JJ, Kohl A, Donald CL (2017) Inhibition of type I interferon induction and signalling by mosquito-borne flaviviruses. Cellular Microbiology 19: e12737.
- Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL (2006) Blocking of Interferon-Induced Jak-Stat Signaling by Japanese Encephalitis Virus NS5 through a Protein Tyrosine Phosphatase-Mediated Mechanism. Journal of Virology 80: 5908-5918.
- Thurmond S, Wang W, Song J, Hai R (2018) Suppression of Type I Interferon Signaling by Flavivirus NS5. Viruses 10: 712.
Citation: Balzan M (2021) Arthropod-Borne Disease May Lead to a More Resilient Type I Interferon Response to Coronavirus by Reducing Genetic Defects of Innate Immunity by Natural Selection. J Clin Immunol Immunother 7: 067.
Copyright: © 2021 Martin Balzan, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

