
Classification of Substances Combining Standoff Laser Induced Fluorescence and Machine Learning
*Corresponding Author(s):
Marian KrausGerman Aerospace Center Dlr, Institute Of Technical Physics, Atmospheric Propagation And Effect, 74239 Lampolds-hausen, Germany
Tel:+49 6298 28339,
Email:marian.kraus@dlr.de
Abstract
Contaminated objects and areas must be handled carefully depending on the underlying pollution. There are methods which require short distances, others the collection of samples or even direct contact to the hazardous, and some of the established techniques take long to reach a conclusion. A fast standoff method for predicting the potential hazard can be achieved by examining the laser induced fluorescence spectra of the substances of interest. The samples are excited by low-energy laser pulses of two alternating wavelengths. The datasets are measured for almost 50 agents, including fuels, pesticides and bacteria and represent the basis for a subsequent classification procedure.
Therefore, the investigated materials are grouped in seven classes depending on their origin and utilization. The majority of the dataset is used in a training phase to create predictive models, which are tested with the remaining signals to qualify the classification. After all, the single spectra of the test set are classified with an error rate less than 01. % in predicting the correct class. With a statement like this first responders would be able to choose the right preventive measure for a rescue or decontamination procedure.
Keywords
INTRODUCTION
Machine learning is one of the keys for online detection of critical substances. For individual experimental setups different methods can be used but they have to be trained with the respective materials of interest. This paper describes a setup for the detection of laser induced fluorescence (LIF) signals as published in [1] as well as the part of data analysis leading to accurate classification models which could be used in online applications making a prediction about the kind of pollution.
For data acquisition a spectral resolution of about 13 nm has been chosen which is sufficient for the analysis of LIF spectra, as indicated in [2] where the classification results of two setups with different spectral resolutions and three different model types are compared. A description of the classification performance for the other setup has been published previously [3]. Further information about online detection techniques like LIF, IR (Infrared) spectroscopy, LIDAR (Light Detection and Ranging) and DIAL (Differential Absorption LIDAR) can be found in [4-7]. Among the different algorithms the signals are classified with a method known as random forests being robust and able to handle large data [8].
MATERIALS AND METHODS
| Bacteria (PBS) | Fuel (pure) | Lubricant (pure) | Marker (water) | Pesticide (water / diethyl ether) | Plant (water) | Solvent (pure) |
| B.atrophaeus | Diesel | Anderol555 | Anthranilic Acid | Isoproturon (d) | Pop.Deltoides | Benzaldehyd |
| B.brevis | Jet fuel | Coconut oil | β-Carotene | Isoproturon (d) | Pop.tremula | Cyclopentan |
| B.fungorum | Kerosene | Colza oil | Chlorophylla | Malathion (w) | Bee pollen spring | Diethyl ether |
| B.pyrrocinia | Paraffin | Motor oil | Isoadenin | Oxyfluorfen (d) | Bee pollen summer | D-Limonene |
| B.subtilis | Pumpkin oil | Lutein | Permethrin (d) | Ethyl Acetate | ||
| B.thuringiensis | Sunflower oil | Lycopene | Terbuthylazine (d) | Isopropyl alcohol | ||
| E.coli | Piperine | Losin100 | ||||
| M.luteus | p-Xylol | |||||
| O.urethralis | Turpentinesubstitute | |||||
| P.fluorescens | ||||||
| P.polymyxa | ||||||
| Y.aldovae |
Table 1: Components of each class plus information about solvents; w=water, d=diethyl ether.
EXPERIMENTAL SETUP
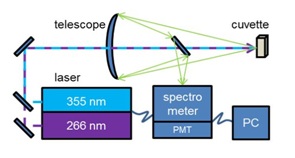
Figure 1: Simplified schematic view of the experimental setup.
DATA PREPROCESS:
First of all, those channels were eliminated which contain no or misleading information: the lower regions beyond both excitation wavelengths and the range of possible Raman peaks whose intensity could influence the scaling process and might lead to misclassifications caused by the presence of different solvents. Due to comparability reasons of different measurements the data were scaled by setting the minimum to 0 and the maximum to 1. The median spectrum of each substance is visualized by one plot per class in figure 2.
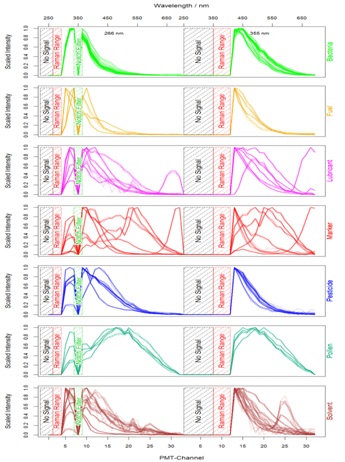 Figure 2: Each plot shows the median of 100 signals for the belonging substances.
Figure 2: Each plot shows the median of 100 signals for the belonging substances.Finally, after modification the data contain information about the class to which they belong to and the normalized signal intensities from 47 features (27 for 266 nm excitation and 20 for 355 nm excitation). This dataset was passed to the model generation process where cross-validation and boot strapping were additionally performed to verify and optimize the classification models [8]. Taking the median (or mean) of several spectra is an optional step to gain even better results in less runtime but is not necessary within this scope and only done for visualization.
A scatter plot of the data of two channels is displayed in figure 3, reduced to the median of five consecutive signals and colored by class for a clearer segmentation. Despite that, the clusters are overlapping and cannot be separated well from each other but the classification is based on using all of the present features.
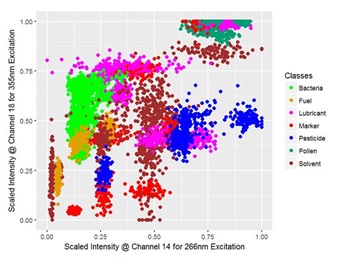 Figure 3: Scatter plot for the scaled signal intensities of two features corresponding to the PMT-channel 14 and 18 which detect a 15 nm broad spectral region around 436.7 nm and 491.4 nm of the fluorescence signal excited by radiation with a wavelength of 266 nm and 355 nm, respectively; for visualization the dataset is reduced to the median of five consecutive spectra and colored by class.
Figure 3: Scatter plot for the scaled signal intensities of two features corresponding to the PMT-channel 14 and 18 which detect a 15 nm broad spectral region around 436.7 nm and 491.4 nm of the fluorescence signal excited by radiation with a wavelength of 266 nm and 355 nm, respectively; for visualization the dataset is reduced to the median of five consecutive spectra and colored by class.CLASSIFICATION
Most of the algorithms provide a few variables which are adjusted during the training phase to gain best possible results. Every combination of those parameters is run separately and the final model can be chosen as the one with the highest accuracy, which is the ratio of the correctly predicted values to the total number of predictions. Within the method random forests many randomly generated decision trees classify the data [14]. The creation of these trees considers basic information of possible codomains of the thresholds and the sets are sampled among the given features. One variable is the amount of trees and another one is the number of sampled features which are randomly selected. The most frequent results of the different models are chosen as the best fit and used to build the final model by averaging their splitting thresholds. This can be processed several times with resampled training sets but within the present classification the tree structure did not change significantly after a few runs.
Instead of grouping the agents in seven classes it is also possible to identify the substances by their LIF spectra within the investigated dataset. Therefore, the modeling process is run again resulting in a new tree which has been trained with that intention. This second model might be used as well for a grouped prediction but it is rather overfitted and worse for that issue.
RESULTS
The predictions are listed in table 2 showing that the utilization of a single excitation wavelength provides enough information to distinguish the classes (99.4 resp. 87.4 %). However using the complete dataset leads to even better results (99.9 %). If the substances are not divided in groups and ought to be discriminated precisely, the effect is more obvious. The single substances can be identified using both spectra with an accuracy of 94.1 % instead of 87.8 % resp. 74.5 % if only one of them is used. Even the very similar spectra of bacteria can be separated as shown in [1].
| 266 nm | |||||||
| Reference | Bacteria | Fuel | Lubricant | Marker | Pesticide | Pollen | Solvent |
| Prediction | |||||||
| Bacteria | 1497 | 0 | 7 | 0 | 0 | 0 | 0 |
| Fuel | 0 | 500 | 0 | 0 | 0 | 0 | 0 |
| Lubricant | 3 | 0 | 726 | 0 | 1 | 0 | 1 |
| Marker | 0 | 0 | 0 | 875 | 0 | 0 | 0 |
| Pesticide | 0 | 0 | 0 | 0 | 746 | 0 | 2 |
| Pollen | 0 | 0 | 0 | 0 | 0 | 500 | 0 |
| Solvent | 0 | 0 | 17 | 0 | 3 | 0 | 1122 |
| Accuracy: 99.4 % (Discrimination: 87.8 %) | |||||||
|
355 nm |
|||||||
| Reference | Bacteria | Fuel | Lubricant | Marker | Pesticide | Pollen | Solvent |
| Prediction | |||||||
| Bacteria | 1394 | 10 | 58 | 0 | 74 | 0 | 92 |
| Fuel | 7 | 381 | 15 | 2 | 46 | 0 | 8 |
| Lubricant | 17 | 14 | 622 | 0 | 12 | 2 | 2 |
| Marker | 2 | 7 | 0 | 835 | 3 | 4 | 20 |
| Pesticide | 41 | 72 | 51 | 2 | 566 | 0 | 49 |
| Pollen | 0 | 0 | 3 | 4 | 0 | 493 | 0 |
| Solvent | 39 | 16 | 1 | 32 | 49 | 1 | 954 |
| Accuracy: 87.4 % (Discrimination: 74.5 %) | |||||||
| 266 & 355 nm | |||||||
| Reference | Bacteria | Fuel | Lubricant | Marker | Pesticide | Pollen | Solvent |
| Prediction | |||||||
| Bacteria | 1498 | 0 | 1 | 0 | 0 | 0 | 0 |
| Fuel | 0 | 500 | 0 | 0 | 0 | 0 | 0 |
| Lubricant | 2 | 0 | 749 | 0 | 0 | 0 | 0 |
| Marker | 0 | 0 | 0 | 875 | 0 | 0 | 0 |
| Pesticide | 0 | 0 | 0 | 0 | 748 | 0 | 0 |
| Pollen | 0 | 0 | 0 | 0 | 0 | 500 | 0 |
| Solvent | 0 | 0 | 0 | 0 | 2 | 0 | 1125 |
Table 2: Confusion matrices for single wavelengths and their combination; the correctly classified spectra are on the main diagonals; also including the accuracies for individually discriminated samples..
DISCUSSION
Additional excitation of other fluorophores like phenylalanine, a compound of living organisms like bacteria, can be achieved using radiation further in the UV spectral region and may lead to an expanded variety of the signals followed by a better classification performance. This is promising especially for the discrimination of bacteria where the signals are dependent on the surroundings and even varies in different growth phases as shown in [15]. Another aspect which has to be investigated is the effect of different mixtures, concentrations and backgrounds and how their impact can be handled with data analysis.
In this paper we present LIF measurements combined with a subsequent classification of 48 different samples. The high accuracy of 99.9 % for the classification and 94.1 % for the identification within the used dataset indicate that a detection system utilizing a comparable model could be able to distinguish different classes of materials. Future measurements will be performed on our free 130 m long transmission test range operated by the DLR in Lampoldshausen, Germany, to investigate atmospheric influences. Furthermore, the sensitivity and the reproducibility of these measurements will be evaluated as well as the impact of spectral changes due to substance concentration variations and solvent effects on the classification.
REFERENCES
- Gebert F, Kraus M, Fellner L, Arne W, Pargmann C, et al. (2018) Novel standoff detection system for the classification of chemical and biological hazardous substances combining temporal and spectral laser induced fluorescence techniques. 1st Scientific International Conference on CBRNe - SICC 2017, Rome, Italy.
- Kraus M, Fellner L, Gebert F, Grünewald K, Pargmann C, et al. (2017) Comparison of Classification methods for Spectral Data of Laser Induced Fluorescence. 1st Scientific International Conference on CBRNe - SICC 2017, Rome, Italy.
- Fischbach T, Duschek F, Hausmann A, Pargmann C, Aleksejev V, et al. (2015) Standoff detection and classification procedure for bioorganic compounds by hyperspectral laser-induced fluorescence. Chemical, Biological, Radiological, Nuclear, and Explosives (CBRNE) Sensing XVI, SPIE, USA. 9455: 9.
- Gaudio P, Gelfusa M, Murari A, Pizzoferrato R, Carestia M, et al. (2017) Application of optical techniques to detect chemical and biological agents. Defence S&T Technical Bulletin 10: 1-13.
- Buteau S, Ho J, Lahaie P, Rowsell S, Simard JR, et al. (2010) Laser based standoff detection of biological agents.
- Hay KG, Norberg O, Normand E, Önnerud H, Black P (2017) Development of an open-path gas analyser for plume detection in security applications. Advanced Optical Technologies 6: 67-73.
- Sun J, Ding J, Liu N, Yang G, Li J (2018) Detection of multiple chemicals based on external cavity quantum cascade laser spectroscopy. Spectrochim Acta A Mol Biomol Spectrosc 191: 532-538.
- Lantz B (2015) Machine Learning with R (2ndedn). Packt Publishing, Birmingham, UK.
- Duschek F, Fellner L, Gebert F, Grünewald K, Köhntopp A, et al. (2017) Standoff Detection and Classification of Bacteria by Multispectral Laser-Induced Fluorescence. Advanced Optical Technologies 6: 75-83.
- RStudio (2018) RStudio: Integrated Development Environment for R. RStudio, Boston, Massachusetts, USA.
- R Development Core Team (2016) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
- Kuhn M (2017) caret: Classification and Regression Training. ASCL, USA.
- Kuhn M, Johnson K (2013) Applied Predictive Modeling. Springer, New York, USA. Pg no: 600.
- Ho TK (1995) Random decision forests: ICDAR '95 Proceedings of the Third International Conference on Document Analysis and Recognition (Vol1). IEEE, Washington, New Jersey, USA. Pg no: 278.
- Fellner L, Gebert F, Walter A, Grünewald K, Duschek F (2017) Fluorescence Spectra of a Bacterial Population During Different Growth Phases. 1st Scientific International Conference on CBRNE, University of Rome Tor Vergata, Rom, Italy.
Citation: Kraus M, Fellner L, Gebert F, Pargmann C, Walter A, et al. (2018) Classification of Substances Combining Standoff Laser Induced Fluorescence and Machine Learning. J Light Laser Curr Trends 1: 003.
Copyright: © 2018 Marian Kraus, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

