
Comparative Gene Expression Profiling and Character Development In stable Angina Pectoris Disease
*Corresponding Author(s):
Fei TengThe Beijing Genomics Institute (BGI), Beishan Industrial Zone, Yantian District, Shenzhen, Guangdong, China
Tel:+86 13632838745,
Email:Sky.teng@bgi.com
Zhong Wang
Institute Of Basic Research In Clinical Medicine, China Academy Of Chinese Medical Sciences, Nanxiaojie, Dongzhimennei, Dongcheng District, Beijing, China
Tel:+86 13601315446,
Email:zhonw@vip.sina.com
Abstract
Objective: To investigate the characteristics and find more effective biomarkers at gene expression level of Stable Angina (SA) pectoris.
Methods: Here blood samples from 45 angina pectoris patients and 20 healthy controls were collected and produced by next generation sequencing and then compared gene expression levels between these two groups to find the significant regulated genes. Several methods or software were used to identify the features of angina pectoris relative to healthy people at transcriptase level.
Results: 874 differential expressed genes may likely to be the candidate biomarkers for angina pectoris patients, 459 genes can be searched from the database Online Mendelian Inheritance in Man (OMIM) which providing information about the human genes and genetic disorders especially pay attention to genetic variants and gene expression.
Discussion: Several different expressed genes are associated with angina pectoris related signal pathways. Such as PDGFA and KDR which play a role in atherosclerosis and already as a target for potential treatment. The function of rest 323 genes still needs to continue the function study in disease. These DEGs can act as candidate biomarkers for diagnose or treat, but still need further validate, refine and extend the process and function in coronary heart disease angina. The identification of biomarkers can be applied to the diagnosis, prognosis, and treatment of stable angina pectoris, that’s what we are counting on.
Keywords
Biomarkers; Gene expression; Stable angina
Introduction
Previous reports suggest that the most common type of heart disease is Coronary Artery Disease (CAD).Chronic stable angina is a main manifestation in approximately half of all patients who present with coronary heart disease, accounting for more than 80% of the cases worldwide [1]. Recently, some studies have demonstrated that miRNA can be involved in regulation of development of CAD patients by contrasting with controls [2,3]. And genomic mutations like SNP have been reported associated with myocardial infarction through Genome-wide association study [4].
This research aimed to investigate the distinctive mRNA features in the stable angina pectoris, and explore the novel biomarkers for disease diagnosis especially for Chinese population.
Methods
Sample collection
Serum samples from 45 patients with stable angina in 301 Hospital and Xuanwu Hospital China were randomly selected into the trials and twenty healthy volunteers joined as controls. The clinical characteristics of patients like sex/age/hyperlipidemia are summarized in table 1. And patients provided informed consent in accordance with local IRB requirements.
|
Patient cohort (n=45) |
|
|
Diagnosis |
Angina |
|
Median Age (years) |
57.31 |
|
Hyperlipidemia, n (%) |
15 (33.3%) |
|
Female, n (%) |
10 (22.2%) |
|
BMI, n(%) |
|
|
<20 |
0 (0%) |
|
20-25 |
19 (42.2%) |
|
>25 |
26 (57.7%) |
|
CCS angina class, n(%) |
|
|
II |
37 (17.8%) |
|
III |
8 (82.2%) |
Table1: Clinical overview of patient, samples and clinical characteristics.
Library and Sequencing
Total RNA was isolated from the serum samples and was pooled together to construct cDNA Libraries. These libraries were subsequently sequenced throw Illumina HiSeq sequencing with pair-end reads of length 2*100bp according to manufacturer’s instructions as previously described.
Data analysis and Statistics
The widely used software Bowtie2 and RSEM were performed to do the alignments and calculate FPKM value of genes respectively. Normalized mRNA expression data were used to perform the clustering analysis, and only kept the gene which expressed in 60% or more samples which as input for R package ‘Consensus Cluster Plus’[5].
The normalized and log-transformed gene expression data then provided as input to the R package ‘iCluster’[6] and we reduce the dataset to top 6,000 most variable genes which measured by median absolute deviation.
The expressed genes were further analyzed between the groups which separated by clustering analysis, and represented the features of patients with stable angina and healthy respectively. Based on the gene expression level, differentially expressed genes were detected by software ‘DESeq2’ [7] with fold-change lager than 2 and p-value smaller than 0.05.
According to the DEGs, we next performed Gene Ontology (GO) classification and KEGG pathway classification and functional enrichment by using R package ‘phyper’. For each P-value, we correct for multiple comparisons by controlling the False Discovery Ratio. And the terms for which the FDR was not greater than 0.01 were defined as statistically significantly enriched. Meanwhile, online software ‘metascape’ [8] also used to perform the pathway and process enrichment analysis.
Results
Expression and clustering
Throw transcriptome sequencing we identified an averaged 15,747genes for each sample. Based on the expressed RNAs hierarchical, clustering analysis were performed to show the relationship between angina pectoris and healthy. The fingure1 showed great divergence between these two groups although some sample from patient has clustered into another group. The clustering analysis indicated that the expression of genes may be involved in the SA disease progression and required further research (Figure 1).
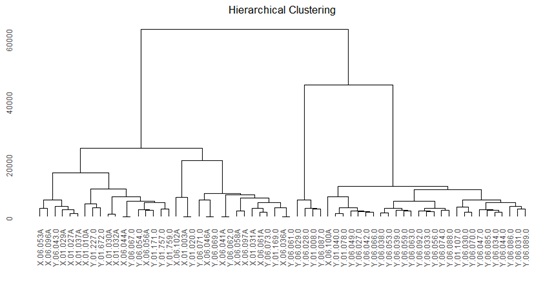 Figure 1: Analysis of hierarchical clustering between samples.
Figure 1: Analysis of hierarchical clustering between samples.
IClusterPlus is developed for integrative clustering analysis of multi-type genomic data, such as DNA copy number alterations and mRNA expressions. In this research, mRNA expression was used as input performing clustering algorithm to discover the subtype of all individuals. Table 2 a indicates the classification results by integrative clustering and figure 2c/d showed the comparing subtypes via PCA. Most samples from healthy are clustered to subgroup 3 and 66.7% of patient samples were divided to subgroup 1. Meanwhile table 2b and figure 2b represented the grouping situations of Consensus Plus were quite similar to result of iClusterPlus.
|
|
ICluster subtypes |
|
|
|
Histology class |
1 |
2 |
3 |
|
Healthy |
0 (0%) |
3 (15%) |
17 (85%) |
|
angina pectoris |
30 (66.7%) |
3 (6.6%) |
12(26.7%) |
|
|
Consensus Cluster |
|
|
|
Histology class |
1 |
2 |
3 |
|
Healthy |
19 (95%) |
1 (5%) |
0 (0%) |
|
angina pectoris |
15 (33.3%) |
22 (48.8%) |
8 (17.7%) |
Table 2: a) Integrated subtype of iCluster. b) Subgroups of Consensus Cluster Plus.
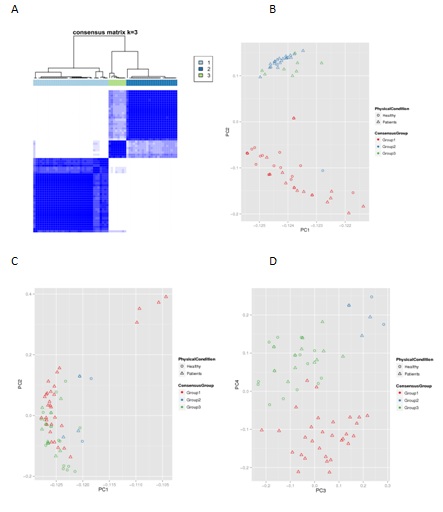 Figure 2: A) The Consensus Matrix plot at k= 3 show a clean consensus distribution of items that have high intra-cluster consensus and low inter-cluster consensus. B) Comparing subtypes via PCA (Consensus Cluster) C) D) Comparing subtypes via PCA (iClusterPlus).
Figure 2: A) The Consensus Matrix plot at k= 3 show a clean consensus distribution of items that have high intra-cluster consensus and low inter-cluster consensus. B) Comparing subtypes via PCA (Consensus Cluster) C) D) Comparing subtypes via PCA (iClusterPlus).
Consensus Cluster Plus is a tool for unsupervised class discovery and Gene expression features were used to detect the possible groups of items. Sample distribution of histology class and consensus cluster is shown for k=3 in table 2b. Percentages are the proportion of the pathology class in a consensus cluster. And found that the angina pectoris and healthy are mainly clustered into separated groups, especially 95% of healthy samples clustered into one group. Both methods showed the clearly divided for healthy and stable angina and can provide guidance for subsequent grouping analysis. The members of each group for the two methods can be found in supplementary table 1. And supplementary figure1 explained why these samples should divide into three groups.
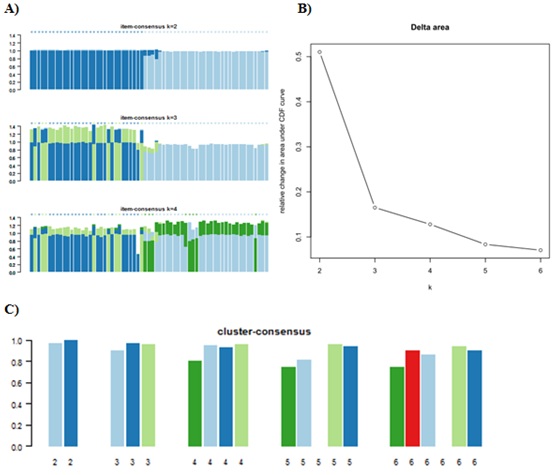 Supplement Figure 1: Graphical plots of Consensus ClusterPlus (A) item-consensus plot. The Item-consensus plot showed that k=3 has items with high consensus to their assigned cluster and low consensus to other clusters denoted by bars of mainly a single color.(B)The Delta Area Plot represents the change in the area under the CDF curves and shows that k=3 is the largest k with an appreciable increase in consensus. (C) cluster-consensus plot. Cluster-consensus (CLC) is the average pair wise IC of items in a consensus cluster. The CLC plot displays these values as a bar plot that are grouped at each k. According to the sum of this evidence, a cluster count of 3 is suite for this project.
Supplement Figure 1: Graphical plots of Consensus ClusterPlus (A) item-consensus plot. The Item-consensus plot showed that k=3 has items with high consensus to their assigned cluster and low consensus to other clusters denoted by bars of mainly a single color.(B)The Delta Area Plot represents the change in the area under the CDF curves and shows that k=3 is the largest k with an appreciable increase in consensus. (C) cluster-consensus plot. Cluster-consensus (CLC) is the average pair wise IC of items in a consensus cluster. The CLC plot displays these values as a bar plot that are grouped at each k. According to the sum of this evidence, a cluster count of 3 is suite for this project.
Differential expressed genes
Differential expression analysis between compared groups was performed to identify the candidate novel biomarkers of angina pectoris. Based on the gene expression level, R packages DESeq2were used to detect the differentially expression genes.
Although it’s hard to find the significant biomarkers while comparing all the healthy and patients since some patient samples mixed in the healthy samples through the clustering. Owe performed the differential analysis for features based on the clustering results while samples from iCluster subtype 1 (subtype2 and subtype3 of consensus) as treatment and all healthy samples as control.
In total874 genes were detected as significant differentially expressed in SA and among them 569 were up-regulated and 305 were down-regulated. It is obvious that a large proportion of genes were over expressed through figure 3 A/B. And based on the gene expression level distribution which showed in figure 3C and listed in table 3, the FPKM value of mainly over expressed genes are larger than 1 in SA group which indicate highly expressed genes may play the leading role in disease. As can be seen from the supplementary figure 2, most differential expressed genes were located in chromosome 1 and 2 and 19 and 12,the function or other meaning was unclear and called for further research.
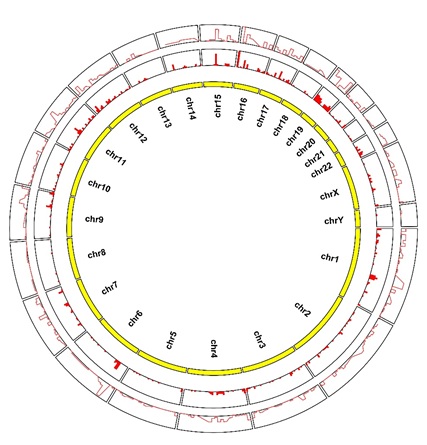 Supplement Figure 2: The chromosome position of different expressed genes.
Supplement Figure 2: The chromosome position of different expressed genes.
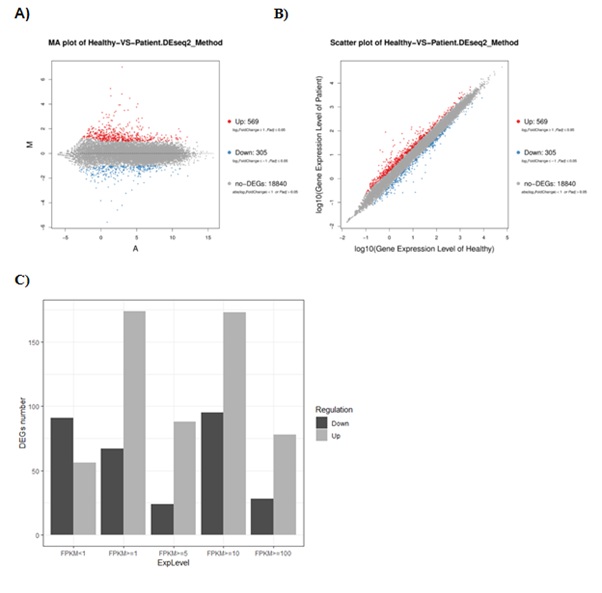 Figure 3: A) MA plot and B) Scatter plot of DEGs. C) Expressed quantity distribution of the DEGs.
Figure 3: A) MA plot and B) Scatter plot of DEGs. C) Expressed quantity distribution of the DEGs.
|
Corhot |
Regulates |
Expression Level |
Counts |
|
Healthy(20) |
Up Regulated (569) |
FPKM<1 |
56 |
|
FPKM>=1 |
174 |
||
|
FPKM>=5 |
88 |
||
|
FPKM>=10 |
173 |
||
|
FPKM>=100 |
78 |
||
|
Patients(30) |
Down Regulated(305) |
FPKM<1 |
91 |
|
FPKM>=1 |
67 |
||
|
FPKM>=5 |
24 |
||
|
FPKM>=10 |
95 |
||
|
FPKM>=100 |
28 |
Table 3: Statistic of Differently Expressed Genes (DEseq2).
Functional of DEGs
Based on the significant differentially expressed genes and KEGG pathway annotation, Figure 4a below showed the pathway classification and functional information which may disturb by expression changes? Meanwhile, the number of up or down regulated genes in corresponding enrichment pathways are listed in Figure 4b. For each given gene list, pathway and process enrichment analysis has been carried out with the following ontology sources: KEGG Pathway, GO Biological Processes (Supplementary Table 2).
 Figure 4: a) Pathway classification of DEGs. X axis represents the number of regulated genes and the left Y axis represents the functional classification. b) The enrichment pathways of regulation genes. The X axis is the terms of pathway and the Y axis is the number of increased or decreased genes.
Figure 4: a) Pathway classification of DEGs. X axis represents the number of regulated genes and the left Y axis represents the functional classification. b) The enrichment pathways of regulation genes. The X axis is the terms of pathway and the Y axis is the number of increased or decreased genes.
|
Pathway Entry |
Name |
Class |
Gene |
|
map04270 |
Vascular smooth muscle contractio |
Organismal Systems; Circulatory system |
72(ACTG2);2983(GUCY1B1);8399(PLA2G10);83698(CALN1); |
|
map05418 |
Fluid shear stress and atherosclerosis |
Human Diseases; Cardiovascular disease |
3791(KDR);5602(MAPK10);5154(PDGFA);387082(SUMO4) |
|
map04975 |
Fat digestion and absorption |
Organismal Systems; Digestive system |
8399(PLA2G10);26279(PLA2G2D) |
|
map04979 |
Cholesterol metabolism |
Organismal Systems; Digestive system |
6770(STAR);11247(NXPH4) |
|
map04261 |
Adrenergic signaling in cardiomyocytes |
Organismal Systems; Circulatory system |
83698(CALN1);23439(ATP1B4);90993(CREB3L1);85442(KNDC1);786(CACNG1) |
Table 4: Angina pectoris related pathways.
Discussion
In present, atherosclerotic disease is a leading cause of human death in the world, coronary heart disease like stable angina is one type of atherosclerotic disease. Here we found four significant higher expressed genes in SA samples which belong to Fluid shear stress and atherosclerosis pathway. The over expressed gene MAPK10/PDGFA/KDR and SUMO4 which showed in figure 5 can promote the occurrence and development of atherosclerosis and coronary heart diseases in signaling pathways.
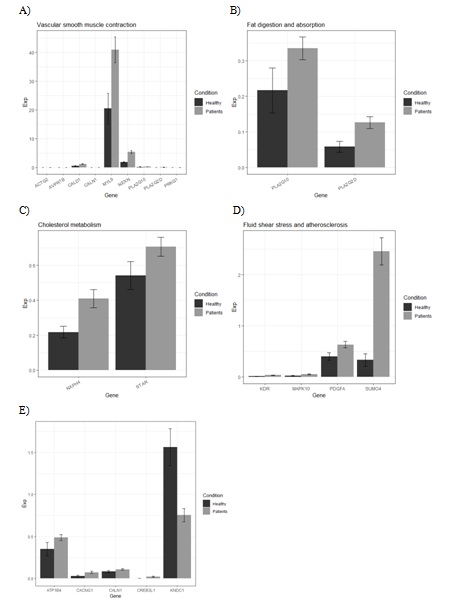 Figure 5: Differential expressed genes in Angina pectoris related pathways. A) Vascular smooth muscle contraction. B) Fat digestion and absorption. C) Cholesterol metabolism. D) Fluid shear stress and atherosclerosis. E) Adrenergic signaling in cardiomyocytes.
Figure 5: Differential expressed genes in Angina pectoris related pathways. A) Vascular smooth muscle contraction. B) Fat digestion and absorption. C) Cholesterol metabolism. D) Fluid shear stress and atherosclerosis. E) Adrenergic signaling in cardiomyocytes.
A member of protein family involved both Platelet-Derived Growth Factors (PDGF) and Vascular Endothelial Growth Factors (VEGF) are encoded by gene PDGFA. Coding preproprotein is proteolytically processing generates subunit A of platelet-derived growth factor, can homodimerize or polymerization of platelet-derived growth factor subunit B. And the proteins and its receptor have been indicated likely to play important roles in multiple developmental processes. Previous scientific work reported that aberrant expressions of PDGF and its receptor (PDGFR) are often be considered as a potential drug target for development of diagnostic or therapeutic agents for multiple disease including atherosclerosis [9]. Inhibition of stromal PDGF receptors can reduced angiogenesis throw depress the expression of PDGFA can be considered a target for potential treatment. And suppressed KDR expression was already as a biomarker of Nilotinib which may have effects on vascular endothelial functions [10].
The main pathogenic factor of atherosclerosis is lipid abnormalities. Hypercholesterolemia can promote atherosclerosis of blood vessels, the most dangerous of which is coronary atherosclerosis, which is the pathogenesis of angina pectoris. The DEGs in Fat digestion and absorption and Cholesterol metabolism should play great roles in the progress of SA disease. Statins are drugs always used to lower cholesterol in treatment of patients with stable angina. Previous research shows active steroid genesis holds an important status in the process of cholesterol movement from outer to the inner mitochondrial via the action of StAR [11]. The observation of disease-related abnormal gene expression also proved the accuracy of this research.
Some other different expressed genes have enriched in the heart-related pathways like adrenergic signaling in cardiomyocyte also have been reported to be related to illness. Previous study has suggested that CALN1 play essential roles in the physiology of neurons and may have a great relationship with learning ability and memory [12] and ATP1B4 may associated with endometriosis pathogenesis. It is also reported that expression of CREB3L1 has correlation with survival time of the glioma patients [13] and the expression of CREB3L1 has been considered as a biomarker for triple negative breast cancer patients who may sensitive to drugs like doxorubicin-based chemotherapy [14]. Previous research has demonstrated that KNDC1could serve as a target for treatment for anti-senescence in the future since knockdown of KNDC1 delays human umbilical vein endothelial cell senescence [15]. These genes might be indirect action with the pathogenesis of angina and need more validation.
Overall, except genes which play its roles in signaling pathways which listed in the table4 there also 459 genes are included in database OMIM which be related to human genes and genetic disorders especially pay attention to genetic variants and gene expression. These genes were already verified or reported the disease related function and some of them play a role in atherosclerosis or already as a target for potential treatment. But still there have 323 genes which not included in databases like KEGG or OMIM which related to healthy or cellular functions. These genes may have some indirect effects and may act as candidate biomarkers for diagnose or treatment still needs further exploration.
Conflict of Interest Statement
Competing Interests: The authors have declared that no competing interests exist.
Data Availability
All the sequencing data are available in China National GeneBank under the accession no.CNP0000461(https://db.cngb.org/search/project/CNP0000461/)
Author’s Contribution
ZW and FT designed and managed the project. Y.W. J.N. and J.G. collect the samples and afford method reference in the evaluation of anticaries. HD and YF performed collection of clinical data. XZ and WQ analyzed the sequencing data and do the statistical. XZ wrote the manuscript, ZW and FT performed the text revision and discussions.
References
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, et al. (2016) Heart disease and stroke statistics-2016 update a report from the American Heart Association. In Circulation 133: 38-360
- Wang X, Lian Y, Wen X, Guo J, Wang Z, et al. (2017) Expression of miR-126 and its potential function in coronary artery disease. African Health Sciences 17: 474-480.
- Cui Y, Song J, Li S, Lee C, Zhang F, et al. (2018) Plasmatic microRNA signatures in elderly people with stable and unstable angina. International Heart Journal 59: 43-50.
- Zhou B, Ho SS, Zhang X, Pattni R, Haraksingh RR, et al. (2018) Whole-genome sequencing analysis of CNV using low-coverage and paired-end strategies is efficient and outperforms array-based CNV analysis. Journal of Medical Genetics 55: 735-743.
- Wilkerson MD, Hayes DN (2010) ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 26: 1572-1573.
- Shen R, Mo Q, Schultz N, Seshan VE, Olshen AB, et al. (2012) Integrative subtype discovery in glioblastoma using iCluster. PLoS ONE 7: 35236.
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15: 1-21.
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, et al. (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10: 1523.
- Torrente D, Cabezas R, Avila M, Sanchez Y, Morales L, et al. (2015) Mechanisms of PDGFRalpha promiscuity and PDGFRbeta specificity in association with PDGFB. Frontiers in Bioscience - Elite 7: 434-446.
- Hadzijusufovic E, Albrecht-Schgoer K, Huber K, Hoermann G, Grebien F, et al. (2017) Nilotinib-induced vasculopathy: Identification of vascular endothelial cells as a primary target site. Leukemia 31: 2388-2397.
- Kraemer FB, Shen WJ, Azhar S (2017). SNAREs and Cholesterol Movement for Steroidogenesis. Mol Cell Endocrinol 441: 17-21.
- Wu YQ, Lin X, Liu CM, Jamrich M, Shaffer LG (2001) Identification of a human brain-specific gene, calneuron 1, a new member of the calmodulin superfamily. Molecular Genetics and Metabolism 72: 343-350.
- Liu LQ, Feng LF, Nan CR, Zhao ZM (2018) CREB3L1 and PTN expressions correlate with prognosis of brain glioma patients. Bioscience Reports 38: 1-8.
- Denard B, Jiang S, Peng Y, Ye J (2018) CREB3L1 as a potential biomarker predicting response of triple negative breast cancer to doxorubicin-based chemotherapy. BMC Cancer 18: 1-7.
- Ji J, Hao Z, Liu H, Liu Y, Liu J, et al. (2018) Effect of KNDC1 overexpression on the senescence of human umbilical vein endothelial cells. Molecular Medicine Reports 17: 7037-7044.
Supplementary Table
Citation: Zhu X, Wang Y, Nan J, Guo J, Dang H, et al. (2021) Comparative Gene Expression Profiling and Character Development In stable Angina Pectoris Disease. J Altern Complement Integr Med 7: 192.
Copyright: © 2021 Xiaolong Zhu, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

