
Journal of Cytology & Tissue Biology Category: Clinical
Type: Short Commentary
DCCL: A Fundamental Dataset of Cervical Cancer Cytological Screen Using Deep Learning Technology
*Corresponding Author(s):
Pifu LuoDepartment Of Pathology, KingMed Diagnostics Co., Ltd., Guangzhou, China
Tel:+86 02028078351,
Email:gz-luopf@kingmed.com.cn
Received Date: Oct 28, 2019
Accepted Date: Nov 19, 2019
Published Date: Nov 26, 2019
ARTICLE
A well-annotated dataset for the Artificial Intelligence (AI)-aided cervical cancer screen, so called Deep Cervical Cytology Lesions (DCCL) has been explored by a collaboration of King Med Diagnostics and Huawei in China. It is the largest set of cervical cytology data for development of the deep learning-based screening product, and it becomes a milestone and “A Benchmark for Cervical Cytology Analysis” as the authors indicated. This beautiful work has been presented at the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI, 2019, Shenzhen, China) [1], and published in the International Workshop on Machine Learning in Medical Imaging [2].
Cervical cancer is one of the most common malignant tumors threatening women's health, especially in the developing countries. It is preventable or curable if its precancerous lesions are early detected by cytological screening combined with Human Papilloma Virus (HPV) test. Due to severely lack of the screening personnel in China, the mortality and morbidity of cervical cancer remain high. An AI-Aided Screening Product (AI-ASP) for cervical cancer detection will be a solution because it helps to screen out the normal cervical specimens so that the cytologists can focus the diagnosis of abnormal lesions.
In the development of an AI-ASP for cervical screening, a large amount of high-quality and annotated cervical cytology dataset is an essential prerequisite for the deep learning algorithm. Lack of dataset for the deep learning training has become a bottleneck of developing any AI-aided product in medicine.
DCCL has collected a total of 14,432 image blocks from 1,167 complete slide images, which is the largest dataset for the deep learning training on cervical cancer screening. These images were selected from a huge volume of cervical pap smears stored in KingMed Diagnostics. KingMed is the largest commercial laboratory in China, and is also the first laboratory in China obtaining Laboratory Accreditation from the College of American Pathologists (CAP) and International Organization for Standardization 15189 (ISO15189). It has accumulated a total volume of 43.5 millions cervical cytological cases over last twenty years. In cervical cytological screening practice, KingMed completely follows the CAP and ISO15189 guidelines in its quality assurance and quality control program. These ensure a high standard resource of DCCL dataset both in quantity and quality.
Figure 1 illustrates the algorithm of DCCL dataset construction. Annotation was performed by eight senior cytopathologists, who have at least six years or above experience of signing-out in cytopathology. Two cytopathologists paired as a group, one does labeling and another does verification. Before the annotation process, cytopathologists were trained by Huawei AI engineers for the labeling standards and lessons to ensure the quality and accuracy of annotation, and to minimize the subjective difference among the cytopathologists. Two types of annotation were provided; one is the slide-level annotation for the normal result and second is cell-level annotation for the abnormal result. A total of 27,972 lesion cells were labeled following the diagnostic criteria and categories of the 2014 Bethesda System (TBS) for the Cervical Cytology Reporting [3]. The annotation results were also randomly checked by a chief cyto pathologist as the quality assurance process. Therefore, the annotation results of DCCL dataset are high quality and reliable.
Cervical cancer is one of the most common malignant tumors threatening women's health, especially in the developing countries. It is preventable or curable if its precancerous lesions are early detected by cytological screening combined with Human Papilloma Virus (HPV) test. Due to severely lack of the screening personnel in China, the mortality and morbidity of cervical cancer remain high. An AI-Aided Screening Product (AI-ASP) for cervical cancer detection will be a solution because it helps to screen out the normal cervical specimens so that the cytologists can focus the diagnosis of abnormal lesions.
In the development of an AI-ASP for cervical screening, a large amount of high-quality and annotated cervical cytology dataset is an essential prerequisite for the deep learning algorithm. Lack of dataset for the deep learning training has become a bottleneck of developing any AI-aided product in medicine.
DCCL has collected a total of 14,432 image blocks from 1,167 complete slide images, which is the largest dataset for the deep learning training on cervical cancer screening. These images were selected from a huge volume of cervical pap smears stored in KingMed Diagnostics. KingMed is the largest commercial laboratory in China, and is also the first laboratory in China obtaining Laboratory Accreditation from the College of American Pathologists (CAP) and International Organization for Standardization 15189 (ISO15189). It has accumulated a total volume of 43.5 millions cervical cytological cases over last twenty years. In cervical cytological screening practice, KingMed completely follows the CAP and ISO15189 guidelines in its quality assurance and quality control program. These ensure a high standard resource of DCCL dataset both in quantity and quality.
Figure 1 illustrates the algorithm of DCCL dataset construction. Annotation was performed by eight senior cytopathologists, who have at least six years or above experience of signing-out in cytopathology. Two cytopathologists paired as a group, one does labeling and another does verification. Before the annotation process, cytopathologists were trained by Huawei AI engineers for the labeling standards and lessons to ensure the quality and accuracy of annotation, and to minimize the subjective difference among the cytopathologists. Two types of annotation were provided; one is the slide-level annotation for the normal result and second is cell-level annotation for the abnormal result. A total of 27,972 lesion cells were labeled following the diagnostic criteria and categories of the 2014 Bethesda System (TBS) for the Cervical Cytology Reporting [3]. The annotation results were also randomly checked by a chief cyto pathologist as the quality assurance process. Therefore, the annotation results of DCCL dataset are high quality and reliable.
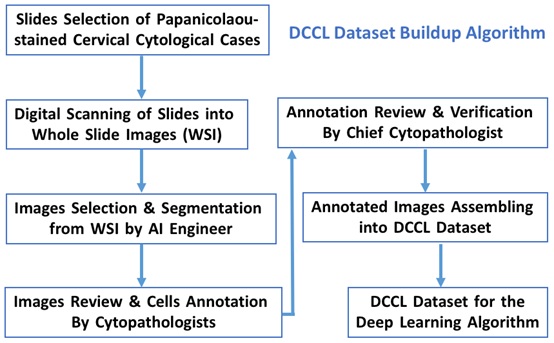 Figure 1: DCCL dataset buildup algorithm.
Figure 1: DCCL dataset buildup algorithm.By using this DCCL dataset, a deep learning algorithm model has developed, which has achieved a sensitivity of 61% of the negative cytological cases signed-out by cytopathologists. In these cases, the accuracy rate is greater than 99% (i.e. less than 1% of false-negative rate). The algorithm has also achieved a 100% of sensitivity of the abnormal cases signed-out by cytopathologists, and no cases were missed among the abnormal ones by the deep learning screening algorithm.
In comparison with the currently available cervical cytological data sets including Cervi SCAN [4], ISBI 2015 [5] and recently published Datasets [6-8] included several hundred of images and few thousands of lesion cells, DCCL dataset has the largest data volume with greater than ten thousands of images and 28 thousands of lesion cells, which come from the largest CAP-accredited laboratory in China. The lesion cell types were classified following TBS criteria [3], and the high-quality dataset was thoroughly labeled and annotated by the highly experienced cytopathologists. It was very time-consuming, laborious and costly process. The dataset will be released and be publically available for the traditional machine learning and deep learning studies. It is very valuable and the blessedness for the development of AI-aided cervical cancer screening.
In comparison with the currently available cervical cytological data sets including Cervi SCAN [4], ISBI 2015 [5] and recently published Datasets [6-8] included several hundred of images and few thousands of lesion cells, DCCL dataset has the largest data volume with greater than ten thousands of images and 28 thousands of lesion cells, which come from the largest CAP-accredited laboratory in China. The lesion cell types were classified following TBS criteria [3], and the high-quality dataset was thoroughly labeled and annotated by the highly experienced cytopathologists. It was very time-consuming, laborious and costly process. The dataset will be released and be publically available for the traditional machine learning and deep learning studies. It is very valuable and the blessedness for the development of AI-aided cervical cancer screening.
REFERENCES
- Zhang CZ, Liu D, Wang LJ, Li YX, Chen XS, et al. (2019) DCCL: A Benchmark for Cervical Cytology Analysis. In: Shen D, Liu T, Peters TM, Staib LH, Essert C, et al. (eds.). Medical Image Computing and Computer Assisted Intervention (MICCAI), Shenzhen, China.
- Zhang CZ, Liu D, Wang LJ, Li YX, Chen XS, et al. (2019) DCCL: A Benchmark for Cervical Cytology Analysis. In: Zhang CZ, Liu D, Wang LJ, Li YX, Chen XS, et al. (eds.). International Workshop on Machine Learning in Medical Imaging, Springer Nature, Switzerland, Pg no: 63-72.
- Nayar R, Wilbur DC (2015) The Pap Test and Bethesda 2014. Acta Cytologica 59: 121-32.
- Tucker JH (1976) CERVISCAN: an image analysis system for experiments in automatic cervical smear prescreening. Comput Biomed Res 9: 93-107.
- https://cs.adelaide.edu.au/simcarneiro/isbi15 challenge/ ?
- Bora K, Chowdhury M, Mahanta LB, Kundu MK, Das AK (2017) Automated classification of Pap smear images to detect cervical dysplasia. Comput Methods Programs Biomed 138: 31-47.
- Zhang L, Le Lu, Nogues I, Summers RM, Liu S, et al. (2017) DeepPap: Deep Convolutional Networks for Cervical Cell Classification. IEEE Journal of Biomedical and Health Informatics 21: 1633-1643.
- William W, Ware A, Basaza-Ejiri AH, Obungoloch J (2019) A pap-smear analysis tool (PAT) for detection of cervical cancer from pap-smear images. Biomed Eng Online 18: 16.
Citation: Che S, Liu D, Zhang C, Tu D, Luo P (2019) DCCL: A Fundamental Dataset of Cervical Cancer Cytological Screen Using Deep Learning Technology. J Cytol Tissue Biol 6: 024.
Copyright: © 2019 Shuanlong Che, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

Journal Highlights
© 2026, Copyrights Herald Scholarly Open Access. All Rights Reserved!
