
Historical Roadmap of Preimplantation Genetic Testing from Past to Present; What is Expected in the Future?
*Corresponding Author(s):
Murat BasarDepartment Of Obstetrics Gynecology And Reproductive Sciences, Yale School Of Medicine, New Haven, CT, United States
Tel:+1 2038238996,
Email:murat.basar@yale.edu
Abstract
Preimplantation Genetic Testing (PGT) has been performed worldwide since 1990 to reveal the sex of the embryos. Until 2010, blastomere biopsy and Fluorescent In Situ Hybridization (FISH) were standard; however, in 2012, trophectoderm biopsy became predominant. Initially, array Comparative Genomic Hybridization (aCGH) was used for analysis and further evolved to Next-Generation Sequencing (NGS), which is used worldwide. Recently non-invasive preimplantation Genetic Testing (ni-PGT) approach arose from the discovery of embryonic DNA in spent embryo culture medium, considering that such methodology would be crucial progress in medically assisted reproduction. In this review, we made a summary of the techniques used in embryo genetic testing from past to present.
Keywords
Aneuploidy; Biopsy; Embryo selection; Non-invasive PGT; Preimplantation genetic testing
Introduction
The goal of Medically Assisted Reproduction (MAR) is to achieve a healthy single birth. Since 1990, various methods have been developed that allow the genetic evaluation of pre-implantation embryos. These techniques have been used to avoid the transmission of a single gene disorder or to select euploid embryos for uterine transfer to increase the chance of ongoing pregnancy rate. All past and current Preimplantation Genetic Testing (PGT) methods have relied upon sampling genetic material from the oocytes/zygotes or embryos/blastocysts. However, the techniques for biopsy are invasive and not entirely risk-free [1]. Additionally, such methods carry costs regarding the need to purchase special equipment and hiring highly skilled embryologists for this technique increases the staffing costs.
Given the complexity consolidated with invasive embryo biopsy technique, it is not surprising that the recent discovery of DNA in embryo culture media, and also in the fluid filling the blastocoel cavity of blastocysts, has ignited excitement about the possibility of non-invasive PGT (niPGT) [2]. Ongoing studies are trying to reveal whether this DNA is a reliable source of information about the embryo's genetic status. If accurate PGT proves possible, less invasive approaches based on 'extra-embryonic' DNA may provide a more straightforward, safer, and cost-effective approach to invasive PGT.
Pre-Implantation Genetic Testing (PGT)
The outcome of chromosome number errors in the post-implantation development is unknown, but they are increasingly recognized as physiological events in the early pre-implantation embryo with many corrective mechanisms in place [3]. To improve clinical outcomes, the increasing precision of screening methods should be used to investigate the effect of such interventions within extensive research initiatives. PGT was first used in Medically Assisted Reproduction (MAR), and it has since become a standard method for examining embryo aneuploidy and genetic disorders in embryos all over the world [4-6] by sifting out embryos with abnormal chromosome numbers. PGT was subdivided into PGT-M (monogenic disorder), PGT-SR (structural rearrangements), which induces miscarriage, and PGT-A (aneuploidy screening). Preimplantation genetic testing-monogenic is restricted to specific gene disorders, whereas preimplantation genetic testing-aneuploidy is a more comprehensive test that looks for aneuploidy in all chromosomes, such as the 22 pairs of autosomal chromosomes and the sex chromosomes X and Y. Preimplantation genetic testing-structural rearrangements are used to screen embryos for chromosome gains and losses caused by parental structural chromosomal abnormalities as translocations, inversions, deletions and insertions [7].
Historically, it was first used by Handyside et al in the United Kingdom to sex embryos in 1990, and the X-linked recessive disease transmissions were prevented by selection of female embryos [8]. In the United States, the first successful PGT for Tay-Sachs disease was performed in 1995 [9]. Until about 2010, cleavage stage embryo biopsy and Fluorescent In Situ Hybridization (FISH) were common; however, Trophectoderm (TE) biopsy became prevalent in 2012 [10]. Furthermore, array Comparative Genomic Hybridization (aCGH) was used for analysis, which evolved into Next Generation Sequencing (NGS), which is now used globally [11].
Pre-implantation genetic testing
Invasive techniques
Even though there are many controversial opinions within these techniques, invasive biopsy methods are still widely used in PGT. Polar Body (PB) biopsy, blastomere biopsy and trophectoderm biopsy can be categorized under invasive techniques. The samples obtained with these techniques have been analyzed by FISH, PCR, aCGH and NGS methods from past to present, and they have made important progress in the development of PGT.
Polar Body (PB) biopsy
Polar body diagnosis (PBD) is an indirect genetic analysis of oocytes. Polar bodies are derivatives of the meiotic cell cycle with no actual impact on embryo development. PBD cannot detect the paternal contribution to the genetic make-up of the developing embryo. The detection of maternally derived chromosomal aneuploidies and translocations in oocytes is the primary application of PBD [12]. The growing interest in PB biopsy and diagnosis has sparked additional research into the technology's versatility. The first and second PB can be removed separately or simultaneously. In the case of fragmentation, it can be difficult to distinguish first PB (PB1) and second PB (PB2) or to accurately separate PB1 and PB2 during a simultaneous biopsy. Recent polarization microscopy studies have revealed that some oocytes with a PB1 may still be in telophase I due to the presence of a connective spindle strand between the PB1 and the oocyte [13]. This type of spindle bridge is meiotic division remnant that occurs during the formation of PB1 as well as at the end of the second meiotic division after the extrusion of the PB2.Too early biopsy increases the risk of spindle remnants in the second PB, while too late biopsy results in a PB1 that has already begun to disintegrate or degenerate [14]. The use of Single-Nucleotide Polymorphism (SNP)-based heterozygosity analysis could aid in the identification and differentiation of PB1 and PB2 [15]. In terms of PB isolation for array-CGH, the timing of PB2 biopsy appears to influence amplification results. It was reported that too early PB2 biopsy may reduce amplification efficiency slightly, but that this effect vanished after adjusting to later biopsy times [16,17].
The interpretation of FISH results with the PB approach is a major issue, particularly for PB1. Whereas PB2 formation and aging can be precisely controlled because the time point of injection initiates the course of PB2 extrusion, this is not the case with PB1.The majority of oocytes are already in metaphase II at the time of ovum pickup, but it is unknown when PB1 was extruded and how long the chromatin of PB1 has been prone to aging [18]. Given the high cost of array-CGH, the economic impact of PB biopsy must also be considered. Because both PBs must be analyzed separately, costs are doubled. With the use of FISH and array-CGH, PB biopsy has been shown [18,19] to be sufficiently effective for the diagnosis of structural and numeric chromosomal abnormalities in human oocytes. Nonetheless, the use of PB biopsy and array-CGH for PGT remains controversial due to cost-effectiveness, the high prevalence of post-meiotic aneuploidies that are undetectable by the PB approach, and questionable precision [20,21].
Blastomere stage biopsy
Blastomere biopsy for PGD is an invasive procedure that involves disrupting cell adhesion and breaching the zona pellucida, then aspirating one or two blastomeres from cleavage-stage embryos. Following the biopsy, the biopsied blastomeres are sent for single cell DNA analysis to screen for a disease-causing mutation and numerous flanking polymorphism markers to help identify the allele (maternal or paternal) that the embryo had inherited. Fluorescence In Situ Hybridization (FISH), one of the PGD technologies, is crucial to its success and is frequently employed to analyze the chromosomal complement in cells. The fundamental idea behind FISH is the homologous complementary hybridization of two single-stranded nucleic acids. To identify and analyze the number and makeup of chromosomes in a single cell, FISH uses fluorescently labeled and chromosome-specific
DNA probes [22].The method caused an earlier compaction and produced signs of developmental delay in both mouse and human embryos, according to studies utilizing static assessment of mouse embryos after blastomere biopsy [23-26]. In early studies, there is a widely cited investigation by Hardy et al., stated that reimplantation development does not appear to be impacted by blastomere biopsy at the eight-cell stage, and although the embryo's metabolism and cell count are both decreased, these changes are proportional to the embryo's decreased cellular mass [27].
Therefore, Scott et al. reported that cleavage-stage biopsies resulted in a considerable reduction in implantation potential [1]. Similarly, according to a study published by Kirkegaard et al., [28] showed that the removal of the blastomeres elongates the period of the precise cell stage at which the biopsy was carried out, allowing the biopsied embryos to advance to succeeding embryonic stages at noticeably later times. As a result, the biopsied embryos' compaction and blastulation times were later than those of the non-biopsied control embryos. To extend the validation of these results, in 2016, Bar-El et al., investigated another study and revealed that blastomere biopsy delays the compaction and the blastulation of the embryos, leading to a decrease in implantation [29]. It is not surprising that Mastenbroek and colleagues noted the failure of PGT when performed by 9-chromosome FISH on biopsied blastomere given the amount of studies demonstrating the ineffectiveness and probable impairment of cleavage stage biopsy in their review and meta-analysis [30,31]. Furthermore, the first and most extensively used method for analyzing PGT-A chromosomes, Fluorescence In Situ Hybridization (FISH), can only evaluate 5 to 12 chromosomes at once [32]. Additionally, signal overlapping, signal splitting, signal diffusion, and probe inefficiency invariably reduced FISH accuracy [33-35].
Trophectoderm (TE) biopsy: PGT v1.0
Since the introduction of the Comprehensive Chromosomal Screening (CCS) technology, TE biopsy for PGT has become a popular treatment option for PGT-A [36]. Kokkali et al., suggested [37] performing embryo biopsy in the blastocyst stage rather than the cleavage stage, when mosaicism is widespread, and few cells may be recovered for chromosomal analysis [38]. Potential for misdiagnosis because of mosaic embryos has been brought up as a concern in PGT. Magli et al., [39] and Josien et al., [40] maintain that the status of aneuploidy and other types of mosaicism do not alter in the inner cell mass or trophectoderms of a blastocyst, despite other studies suggesting that the trophectoderms of blastocysts are frequently present with mosaicism. According to this claim, trophectoderms can be harvested to diagnose the entire embryo (Figure 1). The capacity to extract more cells from a blastocyst is its largest advantage. PGT-A involves the biopsy of 5-6 Trophectoderm (TE) cells (comprising of approximately 3% of total TE cells) from a day-5/6 blastocyst stage embryo [41]. The test requires a sample of between five and eight cells to determine a loss of around 10% of all the cells comprising the blastocyst (about 100-150) [42]. This procedure appears to be substantially less invasive as compared to the cell mass loss indicated by the removal of two blastomeres. The little amount of template DNA makes it challenging to investigate a single blastomere in aCGH and NGS, which are used to examine anomalies in the fundamental structure of the genome. However, because it yields more cells and can be recognized as mosaics, blastocyst biopsy is currently the most popular technique (Table 1).
 Figure 1: Application of Trophectoderm (TE) Biopsy. On the 3rd day of embryo culture, when the embryo cleaved into 8-cells, the zona is hatched by the help of laser. On day 5, trophectoderm cells escapes from the drilled zona to be biopsied (Figure 1A). Biopsy steps of an embryo (Figure 1B).
Figure 1: Application of Trophectoderm (TE) Biopsy. On the 3rd day of embryo culture, when the embryo cleaved into 8-cells, the zona is hatched by the help of laser. On day 5, trophectoderm cells escapes from the drilled zona to be biopsied (Figure 1A). Biopsy steps of an embryo (Figure 1B).
|
Technique |
Year |
Purpose of Performing |
Organism |
Reference |
|
Whole Blastocyst (No Biopsy) |
1967 |
Use of fluorescence microscopy to score sex chromatin and so identify male and female embryos only five days after conception. |
Rabbit |
Gardner et al., Nature, 1967 |
|
Zona Drilling |
1986 |
A micromanipulation apparatus was used to produce holes in the zonae pellucidae of unfertilized mouse oocytes for correction of oligospermia. |
Mouse |
Gordon et al., J Exp Zool, 1986 |
|
Blastomere Biopsy |
1989 |
To develop a technique for micromanipulative removal of a single blastomere from the 4-cell mouse embryo and proliferation of that blastomere in culture. |
Mouse |
Wilton et al., Biology of Reprodution, 1989 |
|
Blastomere Biopsy |
1990 |
To show removal of one or two cells at the 8-cell stage, while reducing the cellular mass, does not adversely affect the preimplantation development of biopsied embryos in vitro and suggest that this approach could be used for |
Human |
Hardy et al., Human Reproduction, 1990 |
|
Blastomere Biopsy |
1990 |
Determine the risk of transmitting recessive X-linked diseases, including X-linked mental retardation, adrenoleukodystrophy, Lesch-Nyhan syndrome and Duchenne Muscular Dystrophy (DMD) |
Human |
Handyside et al., Nature, 1990 |
|
Polar Body (PB) Biopsy |
1990 |
Evaluate the risk for alpha-1-antitrypsin (a-l-AT) deficiency and have transferred only diose embryos derived from oocytes possessing the normal PIM allele. |
Human |
Verlinsky et al., Human Reproduction, 1990 |
|
Blastomere Biopsy & FISH |
1993 |
A short fluorescence in-situ hybridization (FISH) procedure using fluorochrome and digoxigenin labelled DNA probes was developed for application in human preimplantation embryos in order to analyse the five chromosomes most involved in human aneuploidy (X, Y, 18, 13 and 21). |
Human |
Munne et al., Human Reproduction , 1993 |
|
Blastomere Biopsy & WGA |
1994 |
Application of single cell whole genome amplification technology to the analysis of five commonly deleted exons of the dystrophin gene in conjunction with ZFXJ ZFY analysis |
Human |
Kristianson et al., Nature Genetics, 1994 |
|
Blastomere Biopsy & PCR |
1995 |
To determine the ability to apply preimplantation genetic diagnostic techniques to screen for and prevent Tay-Sachs Disease (TSD). |
Human |
Gibbons et al., Fertil Steril, 1995 |
|
Blastomere Biopsy & CGH |
1996 |
To develop a molecular cytogenetic method allowing the simultaneous enumeration of all of the chromosomes in a single cell by using comparative genomic hybridization |
Human |
Wells and Delhanty et al., Eur J Hum Genet, 1996 |
|
Tropectoderm Biopsy & FISH |
1997 |
A new methodology for blastocyst biopsy that uses a 1.48 microm diode laser is described. Trophectoderm cells are biopsied after laster zona drilling and culture, fixed and processed for Fluorescent In Situ Hybridization (FISH) analysis. |
Human |
Veiga et al., Zygote, 1997 |
|
PB1 and PB2 Biopsy & FISH |
1998 |
Genetic testing of oocytes by sampling and Fluorescent In Situ Hybridization (FISH) analysis of the first and second polar bodies, to avoid fertilization and transfer of aneuploid oocytes in IVF patients of advanced maternal age. |
Human |
Verlinsky et al., J Assist Reprod Genet, 1998 |
|
Blastocyst Fluid |
2014 |
To investigate the presence of DNA in Blastocyst Fluids (BFs) and to estimate whether the chromosomal status predicted by its analysis corresponds with the ploidy condition in Trophectoderm (TE) cells, the whole embryo, and that predicted by polar bodies (PBs) or blastomeres. |
Human |
Gianaroli et al., Fertil Steril, 2014 |
|
Morula Biopsy & FISH |
2014 |
To present results from 215 IVF/ICSI cycles with PGS and morula-stage embryos biopsy and to analyze data on percentage of blastocysts, pregnancy rates, birth delivery, and the child’s health status after PGS. |
Human |
Zakharova et al., Plos One, 2014 |
|
Tropectoderm (TE) Biopsy & CCS |
2014 |
To evaluate the relationship between conventional parameters of blastocyst evaluation, comprehensive chromosome screening (CCS) data and implantation potential of euploid blastocysts. |
Human |
Capalbo et al., Human Reproduction, 2014 |
Table 1: A roadmap of invasive biopsy techniques. Historical evolution of invasive biopsy applications.
Quantitative PCR or real-time PCR
A reference gene from the same chromosome is compared to three or four locus-specific amplicons along each chromosome in PCR. This technique can quickly and accurately detect aneuploidy in all 23 pairs of chromosomes although it requires a lot of labor, hence automation is strongly advised [43-45]. While triploidy can be detected by qPCR, structural chromosomal abnormalities are not. Because qPCR does not provide a genotype, it is unable to detect uniparental disomy [45,46].
PGT: v2.0
aCGH
SNPs, or single nucleotide polymorphisms, are extremely variable pairs of single nucleotides (A, T, C, or G) in the genomic DNA of a particular species. The SNPs considered in PGT are typically found in non-exon coding regions of the genome. In PGT, SNP microarrays typically assess over 300,000 SNPs distributed across the genome. A significant role in implantation, miscarriages, or giving birth to a child with a significant genetic abnormality could be played by hundreds of de novo structural chromosome abnormalities that are below the resolution of the 300,000 SNP arrays used for PGT [47-49]. CGH microarrays (aCGH) are less dense than SNP microarrays. CGH arrays are capable of being completed in a shorter timeframe as compared to SNP arrays. Since no genotypes are generated, unlike in SNP arrays, aCGH cannot distinguish between 46, XX and 69, XXX or 46, XY and 69, XXY. Additionally, uniparental disomy cannot be detected by aCGH [50]. All commercial laboratories employ an aCGH for PGT, however it is not developed or validated to detect structural chromosomal abnormalities; it can only detect whole chromosome aneuploidy. aCGH does have a limited ability to detect mosaicism in a trophectoderm sample if the chips are validated against mosaic cell samples [43].
Next-Generation Sequencing (NGS)
Until recently, qPCR and array-based technologies (SNP array and array CGH) were considered the most viable tools for PGT-A as also mentioned above. However, because of the additional data gained and the large cost decrease, NGS has replaced other methods as the preferred method [43]. The reduced chance of miscarriage is among its many benefits and is arguably its most significant one [51]. Alternatively, the identification rate of genetic anomalies by NGS-based PGT-A is high (98%) [52] and it may raise the rate of pregnancies [53]. However, prior to adopting NGS in a clinical diagnostic laboratory, two significant challenges need be addressed: 1. establishing the necessary sequencing settings for each application and the minimal platform resolution to detect deletion/duplication (Del/Dup) and determine the presence of mosaicism; 2. a bioinformatics pipeline and diagnostic algorithms should be developed to minimize the subjectivity associated with the visualization of sequencing plots [54]. The minimum platform resolution, number of reads, and signal/noise ratio all have an impact on the smallest detectable fragment size for del/dup detection [55-57]. The clinical outcome appears to be influenced by the level of mosaicism [58], the identity [59] and the number [60] of affected chromosomes; therefore, a proper validation to define mosaicism thresholds are required for each platform to avoid overdiagnosis due to technical artifacts.
Future total non-invasive approaches: Possible PGT v3.0
In the current gold standard method, the biopsy is required to obtain genetic material for PGD from the developing embryo. The risk of harming embryo development associated with embryo biopsy, the uncertainty of long-term clinical results from biopsy in newborn children, and the high costs associated with the biopsy procedure have prompted researchers working in this field to explore less invasive or non-invasive options and evaluate clinical application possibilities has encouraged [61,62]. For this purpose, some researchers evaluated the approach of aspiration of blastocele fluid (Blastocoel Fluid; BF) with an ICSI needle (Blastocentesis) and examination of the aspirated fluid in terms of showing a less invasive approach [2,63-66]. In other studies, in which the “Spent Culture Media” (SCM) (Figure 2) in which the embryo is grown in a completely non-invasive approach, is used as a source, researchers have shown that the “secretome” features, including nucleic acids, released by the embryo into its environment during development, are successful in euploidy, implantation and clinical success [67-72]. At the current point, the results of the studies suggest that both BF and SCM-based examinations may be sufficient (and more efficient soon) tools for the detection of embryos with high implantation ability (Figure 2); on the other hand, they also point out that they bring many challenges to be overcome when compared to traditional selection and biopsy methods [73]. Considering the disadvantages of the gold standard methods used to determine the implantation potential of embryos today, existing and new studies are progressing towards developing new non-invasive techniques. Applying a non-invasive approach is expected to be beneficial in many ways. It reduces the chance of disrupting/adversely affecting the embryo, its environment, and thus its development potential. Other positive expectations are that similar approaches also eliminate the need for an interventional biopsy, increase their effectiveness and reduce the number of treatments required to achieve a healthy live birth, thus significantly reducing costs. Studies on the evaluation of the embryo's implantation potential with non-invasive approaches can open new perspectives in determining the ploidy state and in selecting the embryo with the highest potential for the live birth outcome. The results of studies performed to date indicate that the approach may be beneficial in terms of “prioritizing” the most suitable embryo for elective Single Embryo Transfer (eSET) in the clinic [74]. In this way, an embryo that can be “selected” with a non-invasive approach without reducing pregnancy success is the most effective way to avoid the problems associated with multiple pregnancies and related adverse medical conditions that may be encountered in both mothers and babies because more than one embryo to be performed with the conventional approach creates pregnancy [75]. Preclinical and clinical studies on non-invasive embryo selection approaches are nowadays called Time-Lapse (TL) morphokinetic studies of embryos in the incubator (which can be combined with artificial intelligence algorithms) using special microscopes and genomic and/or proteomic analysis of the culture media in which the embryo is grown [76,77]. Genomics is the science that studies all functional and structural aspects of the genomes of different species. Examines the whole of genes by applying the techniques of sequencing chromosomes. Metabolomics studies all chemical processes involving metabolites, small molecule substrates, intermediates, and products of cell metabolism. Proteomics, on the other hand, is the large-scale study of proteins and encompasses the investigation of the protein composition, structure and activity level. During the development of non-invasive embryo selection methods, many factors, such as the success of the results to be obtained in preclinical or clinical studies, as well as whether the methodology being developed is user-friendly, whether it can be easily applied in the laboratory and improve MAR results, also determines the widespread use of technology. For instance, in the survey results conducted with the participation of a high number of MAR clinics in the United States and France recently, a significant portion of the participants (57-60%) did not have a TL monitoring system. They knew the techniques mentioned above' superiority over static embryo selection methods. However, it is seen that they plan to have something other than such a system soon [78,79]. These results show that the test or methodology should be easy to adapt and user-friendly and not pose an additional risk in embryo development to ensure the effective use of newly developed or under-development applications that can potentially increase clinical success.
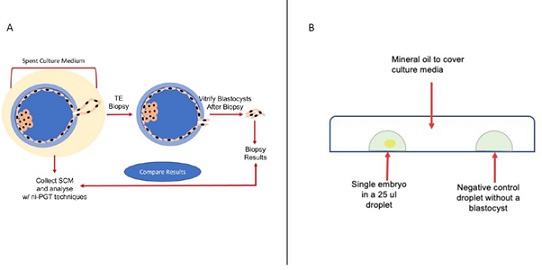 Figure 2: Comparison of two methods as non-invasive and invasive. Recent studies allow the comparison of spent culture media of hatched blastocyst as a non-invasive and TE biopsy results as invasive methods (Figure 2A). Individual blastocysts in individual (25ul) culture media droplets under mineral oil are cultured. Both media droplets as with blastocyst and without blastocyst (as negative control) are collected for further analysis (Figure 2B).
Figure 2: Comparison of two methods as non-invasive and invasive. Recent studies allow the comparison of spent culture media of hatched blastocyst as a non-invasive and TE biopsy results as invasive methods (Figure 2A). Individual blastocysts in individual (25ul) culture media droplets under mineral oil are cultured. Both media droplets as with blastocyst and without blastocyst (as negative control) are collected for further analysis (Figure 2B).
In recent years, basic cell biology studies have examined the relationship between the ploidy state of the cell and its metabolism, and exciting findings have emerged. The study by Rubio et al., is the largest study to date to evaluate embryo ploidy concordance between embryonic cell-free DNA (cfDNA) and TE biopsy. The findings are also consistent with recent publications showing concordance rates between 62.1% and 85.7% [80]. The relationship between the amino acid cycle of the pre-implantation human embryo and its development and pregnancy potential has also been shown in detailed studies. Olcay et al., was studied amino acid turn-over to determine the ploidy state of the embryo non-invasively from the culture fluid, and it was shown that the concentrations of amino acids in the spent embryo culture media differ. Tyrosine (Tyr) level in SCM was found higher in aneuploid group compared to euploid one [81].
Conclusion
Over the years, significant progressions have been introduced in preimplantation genetic testing and medically assisted reproduction, making PGT a well-established, correct, and safe clinical procedure. As sequencing costs continue to decline, PGT moves technically towards a sequencing-based, all-in-one solution for PGT-M, PGT-SR and PGT-A. As patients’ awareness about the risks of transmitting genetic disorders and the number of diseases with identifiable genetic cause(s) continues to rise, the total of treatments and the list of indications for PGT will likely expand. But there is still doubt that embryo biopsy is an invasive technique and needs utmost attention. The recent discovery of DNA in the BF and SCM has generated a surge of interest in the potential for non-invasive PGT. However, the reliability of ni-PGT strategies, concerning the proportion of samples yielding data and the concordance of genetic results relative to those obtained from whole embryos or biopsy specimens, has varied widely between published studies. It is not clear at present which laboratory methods are the most appropriate for the investigation of extra-embryonic DNA. However, as more studies are published a consensus may begin to emerge. Uncertainty over the optimal method for ni-PGT and questions concerning the reliability and clinical utility of the data produced suggest that PGT based upon BCM or SCM samples should, at present, only be carried out in the context of pre-clinical studies and carefully designed clinical pilot investigations.
Disclosure Of Interests
OO, YE, and MB declare no conflict of interests.
References
- Scott RT Jr, Upham KM, Forman EJ, Zhao T, Treff NR (2013) Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril 100: 624-630.
- Palini S, Galluzzi L, De Stefani S, Bianchi M, Wells D, et al. (2013) Genomic DNA in human blastocoele fluid. Reprod Biomed Online 26: 603-610.
- Sciorio R, Dattilo M (2020) PGT-A preimplantation genetic testing for aneuploidies and embryo selection in routine ART cycles: Time to step back? Clin Genet 98: 107-115.
- Dahdouh EM, Balayla J, García-Velasco JA (2015) Comprehensive chromosome screening improves embryo selection: A meta-analysis. Fertil Steril 104: 1503-1512.
- Scott RT Jr, Ferry K, Su J, Tao X, Scott K, et al. (2012) Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: A prospective, blinded, nonselection study. Fertil Steril 97: 870-875.
- Marin D, Xu J, Treff NR (2021) Preimplantation genetic testing for aneuploidy: A review of published blastocyst reanalysis concordance data. Prenat Diagn 41: 545-553.
- Practice Committee of Society for Assisted Reproductive Technology; Practice Committee of American Society for Reproductive Medicine (2008) Preimplantation genetic testing: a Practice Committee opinion. Fertil Steril 90: 136-143.
- Handyside AH, Kontogianni EH, Hardy K, Winston RM (1990) Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature 344: 768-770.
- Gibbons WE, Gitlin SA, Lanzendorf SE, Kaufmann RA, Slotnick RN, et al. (1995) Preimplantation genetic diagnosis for Tay-Sachs disease: Successful pregnancy after pre-embryo biopsy and gene amplification by polymerase chain reaction. Fertil Steril 63: 723-728.
- Takeuchi K (2020) Pre-implantation genetic testing: Past, present, future. Reprod Med Biol 20: 27-40.
- Alyafee Y, Alam Q, Tuwaijri AA, Umair M, Haddad S, et al. (2021) Next-Generation Sequencing-Based Pre-Implantation Genetic Testing for Aneuploidy (PGT-A): First Report from Saudi Arabia. Genes (Basel) 12: 461.
- Montag M, van der Ven K, Rösing B, van der Ven H (2009) Polar body biopsy: A viable alternative to preimplantation genetic diagnosis and screening. Reprod Biomed Online 1: 6-11.
- Montag M, Schimming T, van der Ven H (2006) Spindle imaging in human oocytes: The impact of the meiotic cell cycle. Reprod Biomed Online 12: 442-446.
- De Santis L, Cino I, Rabellotti E, Calzi F, Persico P, et al. (2005) Polar body morphology and spindle imaging as predictors of oocyte quality. Reprod Biomed Online 11: 36-42.
- Treff NR, Scott RT Jr, Su J, Campos J, Stevens J, et al. (2012) Polar body morphology is not predictive of its cell division origin. J Assist Reprod Genet 29: 137-139.
- Magli MC, Montag M, Köster M, Muzi L, Geraedts J, et al. (2011) Polar body array CGH for prediction of the status of the corresponding oocyte. Part II: Technical aspects. Hum Reprod 26: 3181-3185.
- Geraedts J, Montag M, Magli MC, Repping S, Handyside A, et al. (2011) Polar body array CGH for prediction of the status of the corresponding oocyte. Part I: Clinical results. Hum Reprod 26: 3173-3180.
- Montag M, Köster M, Strowitzki T, Toth B (2013) Polar body biopsy. Fertility and Sterility 100: 603-607.
- Montag M, van der Ven K, Dorn C, van der Ven H (2004) Outcome of laser-assisted polar body biopsy and aneuploidy testing. Reprod Biomed Online 9: 425-429.
- Capalbo A, Bono S, Spizzichino L, Biricik A, Baldi M, et al. (2013) Sequential comprehensive chromosome analysis on polar bodies, blastomeres and trophoblast: Insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum Reprod 28: 509-518.
- Treff NR (2013) I11 qPCR-based CCS. Reproductive BioMedicine Online 26: 5.
- Chen CK, Wu D, Yu HT, Lin CY, Wang ML, et al. (2014) Preimplantation genetic diagnosis by fluorescence in situ hybridization of reciprocal and Robertsonian translocations. Taiwan J Obstet Gynecol 53: 48-52.
- Duncan FE, Stein P, Williams CJ, Schultz RM (2009) The effect of blastomere biopsy on preimplantation mouse embryo development and global gene expression. Fertil Steril 91: 1462-1465.
- Ugajin T, Terada Y, Hasegawa H, Velayo CL, Nabeshima H, et al. (2010) Aberrant behavior of mouse embryo development after blastomere biopsy as observed through time-lapse cinematography. Fertil Steril 93: 2723-2728.
- Ajduk A, Zernicka-Goetz M (2013) Quality control of embryo development. Mol Aspects Med 34: 903-918.
- Tarín JJ, Conaghan J, Winston RM, Handyside AH (1992) Human embryo biopsy on the 2nd day after insemination for preimplantation diagnosis: Removal of a quarter of embryo retards cleavage. Fertil Steril 58: 970-976.
- Hardy K, Martin KL, Leese HJ, Winston RM, Handyside AH (1990) Human preimplantation development in vitro is not adversely affected by biopsy at the 8-cell stage. Hum Reprod 5: 708-714.
- Kirkegaard K, Hindkjaer JJ, Ingerslev HJ (2012) Human embryonic development after blastomere removal: A time-lapse analysis. Hum Reprod 27: 97-105.
- Bar-El L, Kalma Y, Malcov M, Schwartz T, Raviv S, et al. (2016) Blastomere biopsy for PGD delays embryo compaction and blastulation: A time-lapse microscopic analysis. J Assist Reprod Genet 33: 1449-1457.
- Mastenbroek S, Twisk M, van der Veen F, Repping S (2011) Preimplantation genetic screening: A systematic review and meta-analysis of RCTs. Hum Reprod Update 17: 454-466.
- Mastenbroek S, Twisk M, van der Veen F, Repping S (2011) Preimplantation genetic screening: A systematic review and meta-analysis of RCTs. Hum Reprod Update 19: 454-66.
- Munné S, Lee A, Rosenwaks Z, Grifo J, Cohen J (1993) Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum Reprod 8: 2185-2191.
- DeUgarte CM, Li M, Surrey M, Danzer H, Hill D, et al. (2008) Accuracy of FISH analysis in predicting chromosomal status in patients undergoing preimplantation genetic diagnosis. Fertil Steril 90: 1049-1054.
- Cohen J, Wells D, Munné S (2007) Removal of 2 cells from cleavage stage embryos is likely to reduce the efficacy of chromosomal tests that are used to enhance implantation rates. Fertil Steril 87: 496-503.
- Shi WH, Jiang ZR, Zhou ZY, Ye MJ, Qin NX, et al. (2021) Different Strategies of Preimplantation Genetic Testing for Aneuploidies in Women of Advanced Maternal Age: A Systematic Review and Meta-Analysis. J Clin Med 10: 3895.
- de Boer KA, Catt JW, Jansen RP, Leigh D, McArthur S (2004) Moving to blastocyst biopsy for preimplantation genetic diagnosis and single embryo transfer at Sydney IVF. Fertil Steril 82: 295-298.
- Cimadomo D, Capalbo A, Ubaldi FM, Scarica C, Palagiano A, et al. (2016) The Impact of Biopsy on Human Embryo Developmental Potential during Preimplantation Genetic Diagnosis. Biomed Res Int 2016: 7193075.
- Kokkali G, Traeger-Synodinos J, Vrettou C, Stavrou D, Jones GM, et al. (2007) Blastocyst biopsy versus cleavage stage biopsy and blastocyst transfer for preimplantation genetic diagnosis of beta-thalassaemia: A pilot study. Hum Reprod 22: 1443-1449.
- Magli MC, Jones GM, Gras L, Gianaroli L, Korman I, et al. (2000) Chromosome mosaicism in day 3 aneuploid embryos that develop to morphologically normal blastocysts in vitro. Hum Reprod 15: 1781-1786.
- Derhaag JG, Coonen E, Bras M, Bergers Janssen JM, Ignoul-Vanvuchelen R, et al. (2003) Chromosomally abnormal cells are not selected for the extra-embryonic compartment of the human preimplantation embryo at the blastocyst stage. Hum Reprod 18: 2565-2574.
- Stern HJ (2014) Preimplantation Genetic Diagnosis: Prenatal Testing for Embryos Finally Achieving Its Potential. J Clin Med 3: 280-309.
- Greco E, Litwicka K, Minasi MG, Cursio E, Greco PF, et al. (2020) Preimplantation Genetic Testing: Where We Are Today. Int J Mol Sci 21: 4381.
- Capalbo A, Treff NR, Cimadomo D, Tao X, Upham K, et al. (2015) Comparison of array comparative genomic hybridization and quantitative real-time PCR-based aneuploidy screening of blastocyst biopsies. Eur J Hum Genet 23: 901-906.
- Brezina PR, Anchan R, Kearns WG (2016) Preimplantation genetic testing for aneuploidy: What technology should you use and what are the differences? J Assist Reprod Genet 33: 823-832.
- Treff NR, Tao X, Ferry KM, Su J, Taylor D, et al. (2012) Development and validation of an accurate quantitative real-time polymerase chain reaction-based assay for human blastocyst comprehensive chromosomal aneuploidy screening. Fertil Steril 97: 819-824.
- Treff NR, Scott RT Jr (2013) Four-hour quantitative real-time polymerase chain reaction-based comprehensive chromosome screening and accumulating evidence of accuracy, safety, predictive value, and clinical efficacy. Fertil Steril 99: 1049-1053.
- Treff NR, Levy B, Su J, Northrop LE, Tao X, et al. (2010) SNP microarray-based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Mol Hum Reprod 16: 583-589.
- Handyside AH (2011) PGD and aneuploidy screening for 24 chromosomes by genome-wide SNP analysis: Seeing the wood and the trees. Reprod Biomed Online 23: 686-691.
- Brezina PR, Kutteh WH (2015) Clinical applications of preimplantation genetic testing. BMJ 350: 7611.
- Chen D, Xu Y, Ding C, Wang Y, Fu Y, et al. (2022) The inconsistency between two major aneuploidy-screening platforms-single-nucleotide polymorphism array and next-generation sequencing-in the detection of embryo mosaicism. BMC Genomics 23: 62.
- Gianaroli L, Magli MC, Ferraretti AP, Fortini D, Grieco N (2003) Pronuclear morphology and chromosomal abnormalities as scoring criteria for embryo selection. Fertil Steril 80: 341-349.
- Simpson JL, Rechitsky S, Kuliev A (2013) Next-generation sequencing for preimplantation genetic diagnosis. Fertility and Sterility 99: 1203-1204.
- Zhang C, Zhang C, Chen S, Yin X, Pan X, et al. (2013) A Single Cell Level Based Method for Copy Number Variation Analysis by Low Coverage Massively Parallel Sequencing. PLoS One 8: 54236.
- García-Pascual CM, Navarro-Sánchez L, Navarro R, Martínez L, Jiménez J, et al. (2020) Optimized NGS Approach for Detection of Aneuploidies and Mosaicism in PGT-A and Imbalances in PGT-SR. Genes 11: 724.
- Hornak M, Horak J, Kubicek D, Navratil R, Tauwinklova G, et al. (2018) The incidence and origin of segmental chromosome abnormalities in human IVF embryos detected during PGD and PGS. Reproductive BioMedicine Online 36: 13-14.
- Popovic M, Dheedene A, Christodoulou C, Taelman J, Dhaenens L, et al. (2018) Chromosomal mosaicism in human blastocysts: the ultimate challenge of preimplantation genetic testing? Hum Reprod 33: 1342-1354.
- Victor AR, Tyndall JC, Brake AJ, Lepkowsky LT, Murphy AE, et al. (2019) One hundred mosaic embryos transferred prospectively in a single clinic: Exploring when and why they result in healthy pregnancies. Fertil Steril 111: 280-293.
- Chuang TH, Hsieh JY, Lee MJ, Lai HH, Hsieh CL, et al. (2018) Concordance between different trophectoderm biopsy sites and the inner cell mass of chromosomal composition measured with a next-generation sequencing platform. Mol Hum Reprod 24: 593-601.
- Grati FR, Gallazzi G, Branca L, Maggi F, Simoni G, et al. (2018) An evidence-based scoring system for prioritizing mosaic aneuploid embryos following preimplantation genetic screening. Reprod Biomed Online 36: 442-449.
- Maxwell SM, Colls P, Hodes-Wertz B, McCulloh DH, McCaffrey C, et al. (2016) Why do euploid embryos miscarry? A case-control study comparing the rate of aneuploidy within presumed euploid embryos that resulted in miscarriage or live birth using next-generation sequencing. Fertil Steril 106: 1414-1419.
- Tocci A (2020) The unknown human trophectoderm: Implication for biopsy at the blastocyst stage. J Assist Reprod Genet 37: 2699-2711.
- Berger JJ (2019) Primum non nocere: are we closer to saying that the trophectoderm biopsy does no harm? Fertil Steril 112: 35-36.
- Gianaroli L, Magli MC, Pomante A, Crivello AM, Cafueri G, et al. (2014) Blastocentesis: A source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil Steril 102: 1692-1699.
- Poli M, Jaroudi S, Sarasa J, Spath K, Child T, et al., The blastocoel fluid as a source of DNA for preimplantation genetic diagnosis and screening. Fertil Steril 100: 37.
- Perloe M, Welch C, Morton P, Venier W, Wells D, et al. (2013) Validation of blastocoele fluid aspiration for preimplantation genetic screening using array Comparative Genomic Hybridization (aCGH). Fertil Steril 100: 208.
- Tobler KJ, Zhao Y, Ross R, Benner AT, Xu X, et al. (2015) Blastocoel fluid from differentiated blastocysts harbors embryonic genomic material capable of a whole-genome deoxyribonucleic acid amplification and comprehensive chromosome microarray analysis. Fertil Steril 104: 418-425.
- Huang G, Zhou C, Wei CJ, Zhao S, Sun F, et al. (2017) Evaluation of in vitro fertilization outcomes using interleukin-8 in culture medium of human preimplantation embryos. Fertil Steril 107: 649-656.
- Capalbo A, Ubaldi FM, Cimadomo D, Noli L, Khalaf Y, et al. (2016) MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil Steril 105: 225-235.
- Katz-Jaffe MG, McReynolds S, Gardner DK, Schoolcraft WB (2009) The role of proteomics in defining the human embryonic secretome. Mol Hum Reprod 15: 271-277.
- Sánchez-Ribas I, Diaz-Gimeno P, Quiñonero A, Ojeda M, Larreategui Z, et al. (2019) NGS Analysis of Human Embryo Culture Media Reveals miRNAs of Extra Embryonic Origin. Reprod Sci 26: 214-222.
- Hammond ER, McGillivray BC, Wicker SM, Peek JC, Shelling AN, et al. (2017) Characterizing nuclear and mitochondrial DNA in spent embryo culture media: Genetic contamination identified. Fertil Steril 107: 220-228.
- Vera-Rodriguez M, Diez-Juan A, Jimenez-Almazan J, Martinez S, Navarro R, et al. (2018) Origin and composition of cell-free DNA in spent medium from human embryo culture during preimplantation development. Hum Reprod 33: 745-756.
- Leaver M, Wells D (2020) Non-invasive Preimplantation Genetic Testing (niPGT): The next revolution in reproductive genetics? Hum Reprod Update 26: 16-42.
- Chen L, Sun Q, Xu J, Fu H, Liu Y, et al. (2021) A Non-invasive Chromosome Screening Strategy for Prioritizing in vitro Fertilization Embryos for Implantation. Front Cell Dev Biol 9: 708322.
- Sullivan EA, Wang YA, Hayward I, Chambers GM, Illingworth P, et al. (2012) Single embryo transfer reduces the risk of perinatal mortality, a population study. Hum Reprod 27: 3609-3615.
- Minasi MG, Greco P, Varricchio MT, Barillari P, Greco E (2020) The clinical use of time-lapse in human-assisted reproduction. Ther Adv Reprod Health 14: 2633494120976921.
- Hernández-Vargas P, Muñoz M, Domínguez F (2020) Identifying biomarkers for predicting successful embryo implantation: Applying single to multi-OMICs to improve reproductive outcomes. Hum Reprod Update 26: 264-301.
- Dolinko AV, Farland LV, Kaser DJ, Missmer SA, Racowsky C (2017) National survey on use of time-lapse imaging systems in IVF laboratories. J Assist Reprod Genet 34: 1167-1172.
- Boueilh T, Reignier A, Barriere P, Freour T (2018) Time-lapse imaging systems in IVF laboratories: A French national survey. J Assist Reprod Genet 35: 2181-2186.
- Rubio C, Navarro-Sánchez L, García-Pascual CM, Ocali O, Cimadomo D, et al. (2020) Multicenter prospective study of concordance between embryonic cell-free DNA and trophectoderm biopsies from 1301 human blastocysts. Am J Obstet Gynecol 223: 751.
- Olcay IO, Akcay B, Bahceci M, Arici A, Boynukalin K, et al. (2022) Noninvasive amino acid turnover predicts human embryo aneuploidy. Gynecol Endocrinol 38: 461-466.
Citation: Olcay O, Ergun Y, Basar M (2023) Historical Roadmap of Preimplantation Genetic Testing from Past to Present; What is Expected in the Future? J Reprod Med Gynecol Obstet 8: 129.
Copyright: © 2023 Orcun Olcay, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

