Multifunctional Role and Regulation of RNAIII of the Agr Quorum Sensing System in Staphylococcus aureus
*Corresponding Author(s):
Ravi Kr GuptaDepartment Of Microbiology And Immunology, University Of Arkansas For Medical Sciences, University Of Arkansas For Medical Sciences, University Of Arkansas For Medical Sciences, Little Rock, United States
Tel:+1 5015267691,
Email:RKGUPTA@uams.edu
Abstract
Advancement of genomics reveals a repertoire of small non-coding RNAs (sRNAs) which are involved in the regulation of many virulence factors of human pathogen such as Staphylococcus aureus. In the past two decades extensive studies on sRNAs in this bacterium confirm their direct or indirect involvement in complex regulatory network. RNAIII of the Agr quorum sensing system is the most extensive studied sRNA in S. aureus. It is the largest known regulatory sRNA of 514 in length with 14 structural stem loops with a half-life of more than 45 min, which suggest that it may, interacts with many targets. A majority of the clinical isolates produce a high level of RNAIII in vitro and in vivo. RNAIII has been shown to play a key role in virulence regulation by affecting many virulence factors. Previous studies suggest that RNAIII directly or indirectly affects many regulatory pathways, virulence factors and global regulators. However, their regulatory mechanisms and its role in regulatory circuits are still not fully understood. This review focus on in-depth analysis of known discoveries of RNAIII regulation, it’s mode of action and future perspectives. The review will help readers to understand the multi factorial role and regulation of RNAIII in complex regulatory circuits of S. aureus.
Keywords
staphylococcus
INTRODUCTION
Small RNAs typically extend from 50 to 600 nts in length and present in order of hundreds in the genome of gram positive and gram negative bacteria. Majority of sRNAs in bacteria were discovered by computational method. They have well defined secondary structures which comprised helix and loop regions for direct and long distance interactions respectively [1,2]. Most of the sRNAs act by blocking the Ribosomal binding site (RBS) of the target mRNA and inhibit the translation but some also bind at the 5’ and 3’ untranslated regions of mRNAs and affect the stability and expression of mRNA [3-5]. Recent advancement in sequencing methods, particularly deep genome sequencing accelerated the small RNA research in bacteria in the last decade. It looks like the central dogma of biology which state the DNA as a master regulator stands to be modified because increasing knowledge of small RNAs suggest that RNA could also regulate DNA and proteins. Staphylococcus aureus is an important human and animal pathogen that causes superficial skin to deep tissue infections [6-8]. The capability of causing a variety of infections by this organism is due to the production of numerous virulence factors which are tightly regulated by transcriptional regulatory proteins, two-component systems and small regulatory RNAs [9-11]. Accessory gene regulator (Agr) is a well-defined quorum sensing system in S. aureus, which plays an important role in virulence gene regulation and affects numerous virulence factors such as capsule polysaccharide, biofilm formation and toxins production [11]. The effector molecule of this two-component system is the largest known small RNA known as RNAIII, which also encodes a small protein, delta hemolysin, at the 5’ region [11]. In the past two decades RNAIII has been extensively studied and well established as a master player in virulence regulation of S. aureus in laboratory as well as clinical strains [12,13]. RNAIII acts via antisense mechanism by direct base pairing with its target mRNAs at post-transcriptional and translational level. It can also affect downstream genes indirectly by targeting transcriptional regulators and proteins [14-17]. This suggests that RNAIII may have numerous targets with diverse mechanisms of regulation in S. aureus. Currently, the known direct targets of RNAIII are hla (alpha hemolysin), spa (protein A), sa1000 (fibrinogen binding protein), sa2353 (coagulase precursor) and transcriptional regulators Rot and mgrA [15-17]. However, Rot and Agr transcriptomes only partially overlap and we have recently shown that RNAIII only affects part of mgrA regulon [17]. This advocates that our understanding of RNAIII mediated virulence regulation in S. aureus is not complete and more research with advance biochemical methods are needed.
RNAIII IS A MEMBER OF QUORUM SENSING SYSTEM IN S. AUREUS
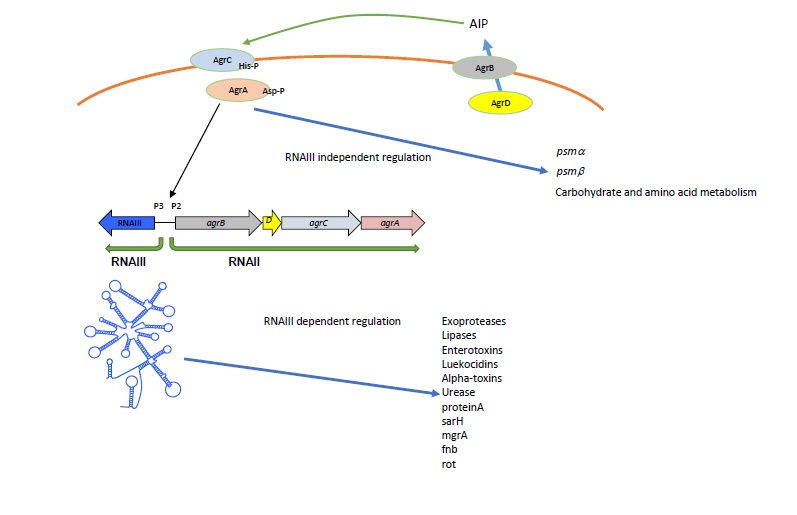
The most well-defined key regulatory systems in S. aureus are gene regulator (Agr) and Staphylococcal accessory regulator (SarA). The Agr quorum sensing system plays a central role in virulence regulation and pathogenesis of S. aureus. Agr regulates the expression of several cell surface proteins, exotoxins, adhesion molecules and virulence factors [9-11]. Its role in pathogenesis has been demonstrated in several animal infection models such a subcutaneous abscess, arthritis and rabbit endocarditis [8-10]. The agr locus generates two primary transcripts, RNAII and RNAIII, from two divergent promoters, P2 and P3, respectively (Figure 1). The P2 operon is a sensory cascade that consist four genes agrB, agrB, agrCand agrA. agrD is a precursor for auto inducing signaling molecule (AIP) which is processed and transported by agrB. agrC is a sensor histidine kinase and agrA is a response regulator. agrC binds to extracellularly accumulated AIP and activates agrA which binds to the promoters P2 and P3 and mediates RNAII and RNAIII transcriptions. RNAIII is a regulatory RNA but also encodes a small toxin called alpha-hemolysin. This positive feedback mechanism of quorum sensing system fine-tunes the regulation of virulence genes at specific cell density [11]. The agr quorum sensing system has two modes of gene regulation. RNAIII independent gene regulation mediated through agrA which includes metabolic and psmgenes, and RNAIII dependent gene regulation which include toxins, proteases, transcription factors and regulatory proteins [18]. RNAIII regulates many genes by direct base pairing with target mRNAs such as hla, spa, fnb, map [12-15] or indirectly through targeting global transcription factors such as Rot and mgrA [12,16,17].
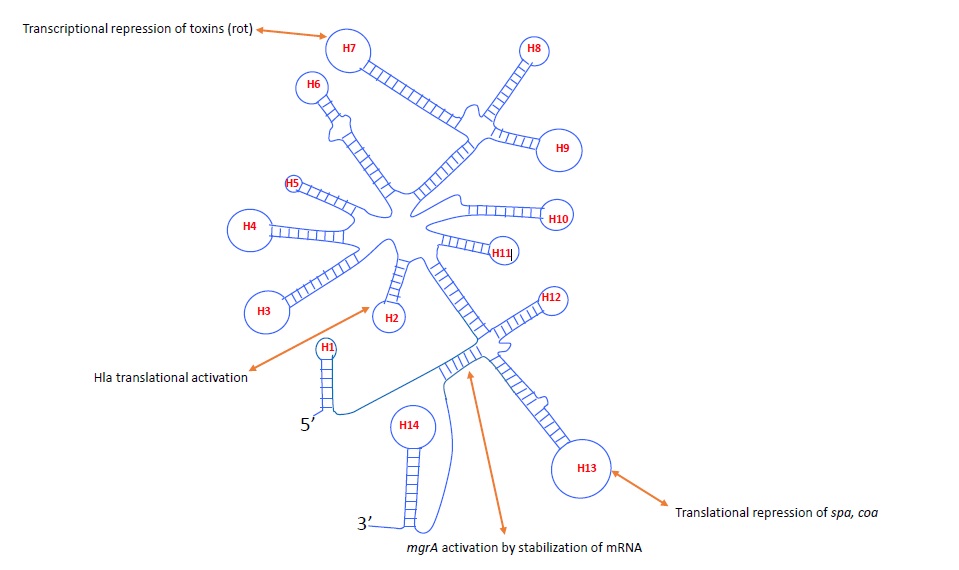
RNAIII IS A MULTI-STRUCTURAL/MULTI-FUNCTIONAL SRNA IN S. AUREUS
Comparative genome analysis of S. aureus strains suggest the presence of 12 sRNAs, of which 7 are present in pathogenicity island (SprA,B,C,D,E,F,G) and 5 in the genome [20]. Another computational approach found 47 novel sRNAs in S. aureus [21,22]. Most of these sRNAs have conserved c-rich motifs which are required for binding at ribosomal binding site and inhibit translation. However, RNAIII which is known as the largest small RNA in S. aureus comprised both short distance and long distance interacting regions (Figure 2). RNAIII is 514 nts in length which consist 14 stem loops and two long distance motif structures [19]. The 5’domain of RNAIII (helix H2 and H3) interacts with alpha-hemolysin mRNA to activate its expression [23]. The helix H7, H13 and H14 has c-rich motifs which interact with target mRNA of rot (repression of toxin), coa (coagulase) and spa (protein A) by antisense mechanism and repress the synthesis of these virulence factors [19] (Figure 2). Rot is a pleiotropic transcription factor which regulates toxins, cell surface proteins, proteases and transporters [24]. RNAIII directly interacts with Rot mRNA posttranscription ally to inhibit protein synthesis [14,15]. Several exoproteases and toxins are repressed by Rot but activated by RNAIII such as serine proteases (splA/F), cysteine proteases (sspA/B), lipase (geh), hemolysins (hla and hld) [15,25]. However, cell surface proteins and virulence factors such as clumping factor B (clfB) and protein A (spa) have been shown to be activated by Rot but repressed by RNAIII [24,26]. Hence, RNAIII and Rot work as antagonist and most of the toxins and cell surface proteins are indirectly regulated by RNAIII through repression of Rot protein [24]. However, transcriptomes of Rot and Agr only partially overlap [12,24]. Several exonucleases such as RexA, DnaQ and cell surface proteins such as clumping factor A, iron regulated cell surface proteins and extracellular matrix and plasma binding proteins are differentially expressed in ?agr but not affected by Rot in our recent RNA-seq analysis (unpublished data). This suggests that RNAIII might interact with other targets that have not been identified. Recently, we reported that RNAIII activates global transcription factor MgrA, which is known to affect more than 300 genes in S. aureus [17,27]. RNAIII directly binds to the 5’ untranslated region of mgrA mRNA and increases its stability to mediate the effect on virulence factors such as capsule polysaccharide, spontaneous autolysis and alpha hemolysin (hla) [17]. However, we found that only part of mgrA regulon are affected through this mechanism. Furthermore, we compared the transcriptomes of Rot, mgrA and Agr and found 64 genes which are differentially expressed in ?agr but not affected by Rot or mgrA [12,24,27]. This emphasizes that our knowledge of RNAIII regulation in S. aureus is still incomplete. Previous studies have linked the Agr system with several two-component systems such as ArlRS, SaeRS, SrrAB [28,29], indicating that RNAIII could interact with these regulators to affect its downstream genes. It is interesting that RNAIII potentially base-pairs with SarT in silico [14], another member of SarA family which repressed the alpha hemolysin [30]. sRNA regulation is majorly through direct RNA-RNA interactions however there are several studies which suggest the indirect regulation. The indirect regulation is mainly mediated through proteins or other RNAs. Several in vitro biochemical methods have been attempted to identify the interacting proteins of a known RNA/sRNA in bacteria [31,32,33]. Recently, an in vitro study using streptavidin aptamer based pull-down assay was used to identify interacting proteins with RNAIII [34]. However, this study lacks the in vivo environment of bacterium which is important for proper folding and secondary structure of RNA and its interactions. RNAIII has been shown to directly interact with a transcriptional regulatory protein WalR [34] and WalR directly bind to the 5’ untranslated region of a major autolysin, lytM in S. aureus [35]. Moreover, microarray study showed that agr repressed the expression of lytM [19]. Thus, RNAIII may indirectly affect the expression of lytM through direct interaction with WalR protein. There are several important regulatory proteins which have been shown to interact with RNAIII in in vitro pull-down assay such as Clp proteases ClpX and ClpC, transcriptional regulator MgrA, WalR and cell division protein FtsZ [34]. These studies suggest that RNAIII interact directly and indirectly with diverse targets including sRNAs, two-component systems, transcriptional regulators and proteins to mediate its regulatory functions.
FUTURE PERSPECTIVES
This review provides a comprehensive overview of RNAIII roles and regulation in S. aureus virulence regulation. We anticipate that availability of super resolution microscopy would help in the real time 3-D structure of RNAIII and its target interactions. High throughput global approaches to identify direct and indirect target molecules will leads to the development of new therapeutic approach against this human and animal pathogen. Our knowledge to establish sRNA as a major cellular component is strengthening day by day by the evident role of sRNAs in cellular processes, regulatory functions and pathogenesis. However, how they fit in the known cellular architecture remain largely unknown. They could function as a mediator of pathway or independently generated and regulate the cellular processes. In present time, antibiotic resistance is a major problem for the available drugs against gram positive as well as gram negative bacteria, therefore development of alternative therapeutic approach is much needed. We believe targeting sRNA could be very useful for drug resistance strains and development of alternative approach to treat infections. It is interesting that sRNA regulation is by transient interaction with its counterpart whether mRNA or protein, hence create less selective pressure on bacteria that suggest targeting sRNA will create less resistance development in bacteria instead of targeting DNA or protein. Now, it is well established that sRNA play significant role in almost all cellular physiology but how bacteria respond to diverse environment and various extracellular and intracellular environment and how these sRNAs integrated in the cellular response will require further studies. Whole genome sequencing and advance computational method will help in further understanding of sRNA regulation in diverse bacterial species.
REFERENCES
- Vogel J, Wagner EG (2007) Target identification of small noncoding RNAs in bacteria. curr Opin Microbiol 10: 262-270.
- Viegas SC, Arraiano CM (2008) Regulating the regulators: How ribonucleases dictate the rules in the control of small non-coding RNAs. RNA Biol 5: 230-243.
- Gupta RK, Luong TT, Lee CY (2015) RNAIII of the Staphylococcus aureus agr system activates global regulator mgrA by stabilizing mRNA. Proc Natl Acad Sci U S A 112: 14036-14041.
- Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615-628.
- Georg J, Hess WR (2011) cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75: 286-300.
- Plata K, Rosato AE, Wegrzyn G (2009) Staphylococcus aureus as an infectious agent: overview of biochemistry and molecular genetics of its pathogenicity. Acta Biochim Pol 56: 597-612.
- David MZ, Daum RS (2010) Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23: 616-687.
- Bunce C, Wheeler L, Reed G, Musser J, Barg N (1992) Murine model of cutaneous infection with gram-positive cocci. Infect Immun 60: 2636-2640.
- Abdelnour A, Arvidson S, Bremell T, Rydén C, Tarkowski A (1993) The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun 61: 3879-3885.
- Cheung AL, Eberhardt KJ, Chung E, Yeaman MR, Sullam PM, et al. (1994) Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Invest 94: 1815-1822.
- Novick RP, Geisinger E (2008) Quorum sensing in Staphylococci. Annu Rev Genet 42: 541-564.
- Dunman PM, Murphy E, Haney S, Palacios D, et al. (2001) Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183: 7341-7353.
- Cassat J, Dunman PM, Murphy E, Projan SJ, Beenken KE, et al. (2006) Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152: 3075-3090.
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, et al. (2007) Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21: 1353-1366.
- Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP (2006) Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61: 1038-1048.
- Liu Y, Mu C, Ying X, Li W, Wu N, et al. (2011) RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. FEBS Lett 585: 899-905.
- Gupta RK, Luong TT, Lee CY (2015) RNAIII of the Staphylococcus aureus agr system activates global regulator mgrA by stabilizing mRNA. Proc Natl Acad Sci U S A 112: 14036-14041.
- Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, et al. (2008) RNAIII-independent target gene control by the agr quorum- sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32: 150-158.
- Benito Y, Kolb FA, Romby P, Lina G, Etienne J, et al. (2000) Probing the structure of RNAIII, the Staphylococcus aureus agrregulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6: 668-679.
- Pichon C, Felden B (2005) Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci U S A, 102: 14249-14254.
- Livny J, Teonadi H, Livny M, Waldor MK (2008) High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS ONE 3: 3197.
- Guillet J, Hallier M, Felden B (2013) Emerging functions for the Staphylococcus aureus PLoS Pathog 9: 1003767.
- Morfeldt E, Taylor D, von Gabain A, Arvidson S (1995) Activation of alphatoxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 14: 4569-4577.
- Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, et al. (2003) Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185: 610-619.
- Oscarsson J, Tegmark-Wisell K, Arvidson S (2006) Coordinated and differential control of aureolysin (aur) and serine protease (sspA) transcription in Staphylococcus aureus by sarA, rot and agr (RNAIII). Int J Med Microbiol 296: 365-380.
- Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, et al. (2005) Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J 24: 824-835.
- Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY (2006) Transcription Profiling of the mgrA Regulon in Staphylococcus aureus. J Bacteriol 188: 1899-1910.
- Yarwood JM, Schlievert PM (2003) Quorum sensing in Staphylococcus J Clin Invest 112: 1620-1625.
- Chabelskaya S, Bordeau V, Felden B (2014) Dual RNA regulatory control of a Staphylococcus aureus virulence factor. Nucleic Acids Res 42: 4847-4858.
- Schmidt KA, Manna AC, Gill S, Cheung AL (2001) SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect Immun 69: 4749-4758.
- Ponchon L, Dardel F (2007) Recombinant RNA technology: the tRNA scaffold. Nat Methods 4: 571-576.
- Iioka H, Loiselle D, Haystead TA, Macara IG (2011) Efficient detection of RNA-protein interactions using tethered RNAs. Nucleic Acids Res 39: 53.
- Jones RC, Deck J, Edmondson RD, Hart ME (2008) Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J Bacteriol 190: 5265-5278.
- Zhang X, Zhu Q, Tian T, Zhao C, Zang J, et al. (2015) Identification of RNAIII-binding proteins in Staphylococcus aureus using tethered RNAs and streptavidin aptamers based pull-down assay. BMC Microbiol 15: 102.
- Dubrac S, Boneca IG, Poupel O, Msadek T (2007) New Insights into the WalK/WalR (YycG/YycF) Essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in.
Citation: Yadav S, Gupta RK (2017) Multifunctional Role and Regulation of RNAIII of the Agr Quorum Sensing System in Staphylococcus aureus. Adv Microb Res 1: 001.
Copyright: © 2017 Shilpi Yadav, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.