
Journal of Plant Science Current Research Category: Agriculture
Type: Review Article
Natural Variation in Wild Gossypium Species as a Tool to Broaden the Genetic Base of Cultivated Cotton
*Corresponding Author(s):
Rosalyn B Angeles-ShimDepartment Of Plant And Soil Science, College Of Agricultural Sciences And Natural Resources, Texas Tech University, Lubbock, Texas, United States
Tel:+1 (806)8346121,
Email:rosalyn.shim@ttu.edu
Received Date: Mar 09, 2018
Accepted Date: May 03, 2018
Published Date: May 18, 2018
Abstract
Cotton is the world’s most important natural textile fiber and a significantly growing source of food stuff, oil and feeds. Among the 53 Gossypium species, only 4 are cultivated, with G. hirsutum and G. barbadensecomprising over 90% of the total cotton cultivation area worldwide. The extensive use of only a few closely related genotypes of cotton, coupled with the widespread adoption of transgenic cultivars, has greatly reduced the genetic base of the crop. This genetic uniformity makes cotton highly vulnerable to emerging biotic and abiotic challenges. Future breeding targets have to seriously consider infusing novel genetic variation into the gene pool of cultivated cotton that can buffer the crop against agro-environmental challenges brought about by shifts in climate. The wild Gossypium species hold a tremendous amount of untapped genetic diversity that can be exploited to broaden the genetic base of cotton. This review highlights the important agronomic traits that have been reported in wild Gossypiumspecies and discusses the various pre-breeding strategies that have been utilized to incorporate genomes of wild Gossypium in cultivated cotton. Genetic and molecular studies towards understanding Verticilliumwilt resistance and salt tolerance in wild cotton relatives are presented in brief.
Keywords
Cotton; Gossypium; Salt tolerance; Verticillium wilt; Wild species
INTRODUCTION
Modern plant breeding has profoundly impacted agricultural production through the development and deployment of varieties with increased yield and improved agronomic performance. However, intensive selection that accompanies contemporary breeding strategies has also introduced a very high degree of genetic uniformity in the field, making crops vulnerable to emerging biotic and abiotic challenges [1]. Crop failures due to the heavy dependence on only a few crop varieties have been documented throughout the history of agriculture. In 1845 for example, a strain of Phytophthora infestans ((Mont.) de Bary) that was accidentally introduced from North America to Ireland decimated the genetically uniform potato varieties cultivated by farmers, leading to the Irish potato famine [2]. In the 1950s, the Panama disease caused by Fusarium oxysporum wiped out the banana variety Gros Michel that was widely cultivated in Central America [3]. The widespread planting of a single corn hybrid variety in the southern US resulted in economic losses of more than a billion dollars in the 1970s due to an outbreak of a new race of the fungal pathogen, Bipolaris maydis [4,5]. In the 1980s, monocultures of a single type of grapevine root forced California grape growers to replant approximately two million acres of vines following an outbreak of a new race of the grape phylloxora, Daktulosphaira vitifoliae [6].
As in the case of most cultivated crops, the overriding emphasis on only a few agronomic traits (i.e., yield and fiber quality) during domestication has severely narrowed the genetic base of cotton. Generations of industry-scale cultivation of a relatively small number of genetically related varieties has further reduced the available genetic variation in this crop. In the US, 98% of cotton that is currently being grown is G. hirsutum [7] and 85% are genetically modified for improved herbicide and pest resistance [8]. The genetic uniformity in cultivated varieties put the cotton industry at a high risk of collapse in the likely event of a disease or pest outbreak or upsurge.
Compounding the threats of disease and pest epidemics are abiotic challenges such as drought, heat and cold, as well as salinization of agricultural lands from years of intensified cultivation. While breeding objectives for cotton remain focused on improving baseline production and product quality, the emerging challenges in agriculture require that new cotton cultivars are developed with adaptation to extremes of temperature, reduced precipitation and saline soils, as well as resistance to new biotypes of pathogens and pests [9]. To this end, cotton breeders need to expand the cultivated germplasm base for the crop and consciously bring in genetic variation from diverse genetic resources that can provide tolerance to a multitude of environmental stresses as well as durable forms of resistance to pests and diseases.
Wild or exotic germplasm constitutes an important resource that can provide novel genetic diversity in cultivated crops that has been lost during domestication. Utilization of naturally occurring genetic variation from wild relatives of crops has been generally perceived as a better option (as opposed to artificial variation) in plant breeding because of the certain selective pressures that has already acted on the fitness of the organism [10]. Wild progenitors of domesticates are commonly found in marginal habitats that are unsuitable for agriculture and that are subject to severe biotic and abiotic stresses. Without human intervention, these wild relatives evolved adaptive mechanisms that allow them to survive harsh environments [10,11]. One such example is Solanum lycopersicoides , a tomato-like nightshade species that thrives in the western slopes of the main Andean cordillera in the Chile-Peru frontier. At 3800 m above sea level, S. lycopersicoides survives frosts and light freezes in well-exposed sites where the cold air from glaciers and snowfields drains. Field cultures of this wild species also exhibit resistance to viral diseases and Lepidopteran pests [12,13]. Yet another example is Hordeum spontaneum, a wild relative of barley. H. spontaneum shares a niche with halophytic vegetation in the Dead Sea coast which receives only a minimum average precipitation of 55 mm per year [14]. The autoecology of S. lycopersicoides and H. spontaneum suggests the presence of considerable genetic variation that lends phenotypic plasticity in both species, allowing them to withstand marginal environments.
The genus Gossypium to which cotton belongs has more than 50 well-established species, only 4 of which are cultivated. In terms of fiber production and quality, the wild cotton relatives are relatively inferior compared to the cultivated species. Despite this, the wild Gossypium germplasm serves as a rich reservoir of novel alleles that can be utilized to improve trait performance in cultivated cotton [7,15]. Interspecific hybridization to broaden the genetic base of the existing cultivars would be an important first step in utilizing the abundant genetic variation from the wild cotton relatives.
This review highlights the useful agronomic traits that have been reported for the different species of wild Gossypium during the past decade. Valuable genetic resources that incorporate the genome of wild Gossypium into the cultivated cotton by conventional breeding or with the aid of biotechnological techniques, as well as the potential application of these resources for trait improvement are discussed. Investigations toward unlocking the genetic and molecular basis of Verticillium wilt resistance and salt tolerance in different wild cotton relatives are presented in brief.
As in the case of most cultivated crops, the overriding emphasis on only a few agronomic traits (i.e., yield and fiber quality) during domestication has severely narrowed the genetic base of cotton. Generations of industry-scale cultivation of a relatively small number of genetically related varieties has further reduced the available genetic variation in this crop. In the US, 98% of cotton that is currently being grown is G. hirsutum [7] and 85% are genetically modified for improved herbicide and pest resistance [8]. The genetic uniformity in cultivated varieties put the cotton industry at a high risk of collapse in the likely event of a disease or pest outbreak or upsurge.
Compounding the threats of disease and pest epidemics are abiotic challenges such as drought, heat and cold, as well as salinization of agricultural lands from years of intensified cultivation. While breeding objectives for cotton remain focused on improving baseline production and product quality, the emerging challenges in agriculture require that new cotton cultivars are developed with adaptation to extremes of temperature, reduced precipitation and saline soils, as well as resistance to new biotypes of pathogens and pests [9]. To this end, cotton breeders need to expand the cultivated germplasm base for the crop and consciously bring in genetic variation from diverse genetic resources that can provide tolerance to a multitude of environmental stresses as well as durable forms of resistance to pests and diseases.
Wild or exotic germplasm constitutes an important resource that can provide novel genetic diversity in cultivated crops that has been lost during domestication. Utilization of naturally occurring genetic variation from wild relatives of crops has been generally perceived as a better option (as opposed to artificial variation) in plant breeding because of the certain selective pressures that has already acted on the fitness of the organism [10]. Wild progenitors of domesticates are commonly found in marginal habitats that are unsuitable for agriculture and that are subject to severe biotic and abiotic stresses. Without human intervention, these wild relatives evolved adaptive mechanisms that allow them to survive harsh environments [10,11]. One such example is Solanum lycopersicoides , a tomato-like nightshade species that thrives in the western slopes of the main Andean cordillera in the Chile-Peru frontier. At 3800 m above sea level, S. lycopersicoides survives frosts and light freezes in well-exposed sites where the cold air from glaciers and snowfields drains. Field cultures of this wild species also exhibit resistance to viral diseases and Lepidopteran pests [12,13]. Yet another example is Hordeum spontaneum, a wild relative of barley. H. spontaneum shares a niche with halophytic vegetation in the Dead Sea coast which receives only a minimum average precipitation of 55 mm per year [14]. The autoecology of S. lycopersicoides and H. spontaneum suggests the presence of considerable genetic variation that lends phenotypic plasticity in both species, allowing them to withstand marginal environments.
The genus Gossypium to which cotton belongs has more than 50 well-established species, only 4 of which are cultivated. In terms of fiber production and quality, the wild cotton relatives are relatively inferior compared to the cultivated species. Despite this, the wild Gossypium germplasm serves as a rich reservoir of novel alleles that can be utilized to improve trait performance in cultivated cotton [7,15]. Interspecific hybridization to broaden the genetic base of the existing cultivars would be an important first step in utilizing the abundant genetic variation from the wild cotton relatives.
This review highlights the useful agronomic traits that have been reported for the different species of wild Gossypium during the past decade. Valuable genetic resources that incorporate the genome of wild Gossypium into the cultivated cotton by conventional breeding or with the aid of biotechnological techniques, as well as the potential application of these resources for trait improvement are discussed. Investigations toward unlocking the genetic and molecular basis of Verticillium wilt resistance and salt tolerance in different wild cotton relatives are presented in brief.
GENETIC DIVERSITY IN THE GENUS GOSSYPIUM
The genus Gossypium includes 46 diploid (2n = 26) and 7 allotetetraploid (2n = 52) species representing the AA, BB, CC, DD, EE, FF, GG, KK and AADD genomes (Table 1).
The genus diverged from its closest relatives, Kokia and Gossypioides, approximately 5-10 million years ago, whereas speciation was estimated to have occurred 1-5 million years ago. Long-distance, transoceanic dispersal was proposed to have driven the evolution of the diploid species, whereas wide hybridization between species having the A and D genomes and subsequent polyploidization gave rise to the allotetraploids (Figure 1) [7,51,52,53]. Species within the genus are geographically distributed in the arid and semi-arid regions of the tropics and sub-tropics, with new exotic species still being discovered in Australia and in the isolated islet chain in the West Pacific [7,28,50,54].
| Wild Species | Useful Agronomic Traits | Origin | Genome |
| G. hirsutum | Widely cultivated | Central America | AD1 |
| G. barbadense | Widely cultivated, long and high quality lint, resistance to Verticillium wilt [16] | South America | AD2 |
| G. tomentosum | Tolerance to heat, source of the nectariless trait for resistance against tarnished plant bug, fleahoppers, boll rot and bollworm [17], resistance to jassids and thrips [18], high fiber quality, fiber length and fiber fineness [19] | Hawaiian Islands | AD3 |
| G. mustelinum | Longer fibers [20,21] | NE Brazil | AD4 |
| G. darwinii | Finer fibers, drought tolerance, resistance to Fusarium and Verticillium wilt [22] | Galapagos Islands | AD5 |
| G. ekmanianum | Dominican Republic | AD6 | |
| G. stephensii | Wake Atoll | AD7 | |
| G. africanum | High fiber strength [23] | Southern Africa | A |
| G. herbaceumL | Resistance to hoppers, white flies, thrips, aphids, and leaf curl virus [24] | Southern Africa | A1 |
| G. arboreumL | Resistance to hoppers, white flies, aphids, leaf curl virus [24], thrips [25], drought tolerance, resistance to black root rot, reniform nematodes [26] and spider-mites [27] | Indus valley, Madagascar | A2 |
| G. anomalum | Resistance to cotton wilt, angular leaf spot, springtails and aphids, drought tolerance, high fiber quality, cytoplasmic male sterility [28] | Africa (Angola, Namibia) | B1 |
| G. triphyllum | Flower color range from blue to purple [29] | Namibia in Africa | B2 |
| G. capitis-viridis | High fiber quality, strong resistance to Verticillium and Fusarium wilt [30] | Cape Verde Islands | B3 |
| G. trifurcatum | Somalia | B | |
| G. sturtianum |
Glandless-seed and glanded-plant [31] Resistance to Fusarium wilt [32] |
Australia | C1 |
| G. nandewarense | Australia | C1N | |
| G. robinsonii | WA, Australia | C2 | |
| G. thurberi | Tolerance to mild frost via defoliation, high resistance to Verticilliumdahlia [29,33] | Mexico | D1 |
| G. armourianum | Resistance to white flies, bacterial blight and jassids [18,29,34] | Mexico | D2-1 |
| G. harknessii | Cytoplasmic male sterility and fertility restorer [35] | Mexico | D2-2 |
| G. davidsonii | Salinity tolerance [36] | Mexico | D3-d |
| G. klotzschianum | Salinity tolerance [37] | Galapagos Islands | D3-k |
| G. aridum |
Salinity tolerance [38] Resistance to reniform nematode [39] |
Mexico | D4 |
| G. raimondii | Resistance to jassids [34] | Peru | D5 |
| G. gossypioides | Resistance to cotton leaf curl disease [40] | Mexico | D6 |
| G. lobatum | Resistance to Verticillium wilt [41] | Mexico | D7 |
| G. trilobum | Cytoplasmic male sterility and restorer factor. Drought tolerance, resistance to bollworm, pink worm, boll rot, Verticillium and Fusarium wilt [29,42] | Western Mexico | D8 |
| G. laxum | Mexico | D9 | |
| G. turneri | Caduceus involucels [43] | Mexico | D10 |
| G. schwendimanii | Mexico | D11 | |
| G. stocksii | Strong fibers, resistance to leaf curl virus [44], resistance to reniform nematode [45] | East Africa, Arabia | E1 |
| G. somalense |
Resistance to reniform nematode [45] Extra fiber strength, resistance to Egyptian bollworm and pink bollworm, arid tolerance [46] |
Northeastern Africa | E2 |
| G. areysianum | Arabia | E3 | |
| G. incanum | Arabia | E4 | |
| G. benadirense | Somalia, Ethiopia, Kenya | E | |
| G. bricchettii | Somalia | E | |
| G. vollesenii | Somalia | E | |
| G. longicalyx | Resistance to reniform nematode [45,47] | Africa | F1 |
| G. bickii | Glandless-seed and glanded-plant [48] | Central Australia | G1 |
| G. australe |
Glandless-seed and glanded-plant, resistance to aphids and spider-mites [49] Resistance to Fusarium and Verticillium wilts, drought tolerance [50] |
Australia | G2 |
| G. nelsonii | Australia | G3 | |
| G. costulatum | Australia | K1 | |
| G. populifolium | WA, Australia | K2 | |
| G. cunninghamii | Northern NT, Australia | K3 | |
| G. pulchellum | WA, Australia | K4 | |
| G. pilosum | WA, Australia | K5 | |
| G. anapoides | Australia | K6 | |
| G. enthyle | WA, Australia | K7 | |
| G. exiguum | WA, Australia | K8 | |
| G. londonderriense | Australia | K9 | |
| G. marchantii | Australia | K10 | |
| G. nobile | WA, Australia | K11 | |
| G. rotundifolium | WA, Australia | K12 |
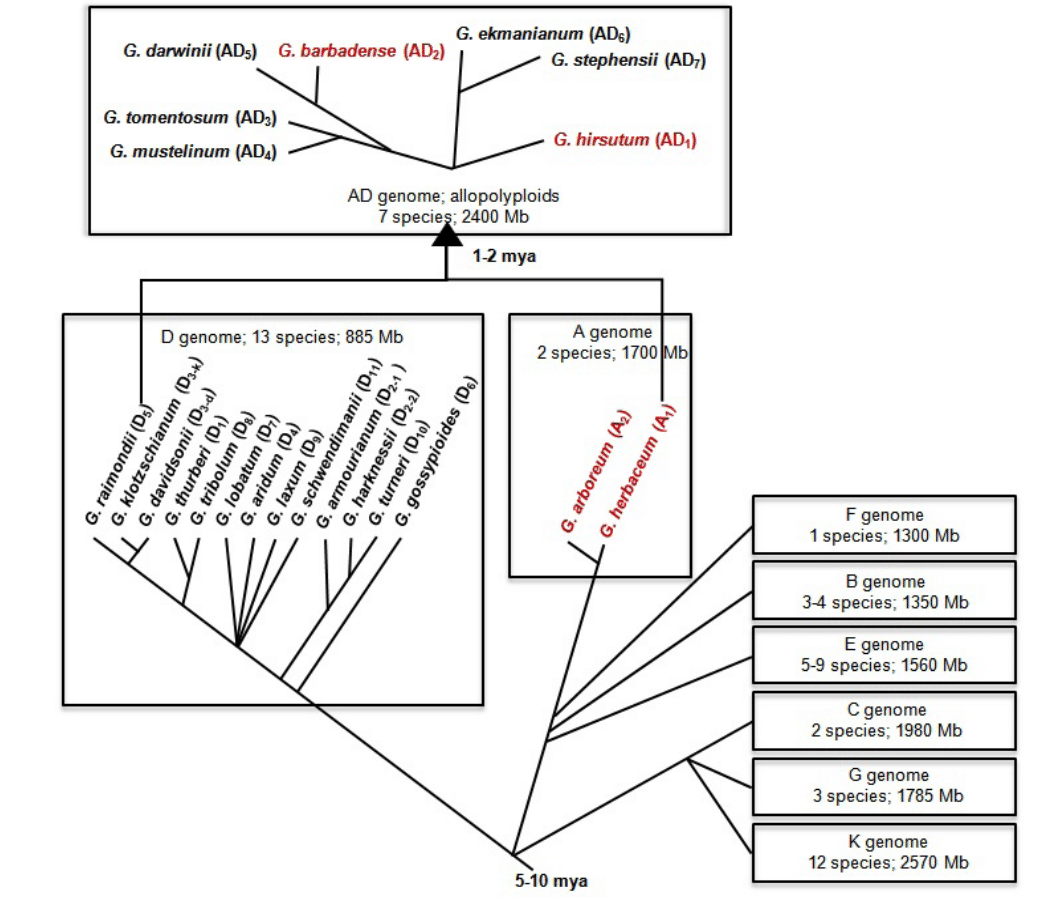 Figure 1: Evolutionary relationship among the different cotton species that evolved after the divergence of Gossypium from the genus Kokia and Gossypioides 5-10 million years ago (adapted from 7,51-54). Letters in parenthesis indicate the genome assignment for each species. Red text indicates cultivated species belonging to the genus Gossypium.
Figure 1: Evolutionary relationship among the different cotton species that evolved after the divergence of Gossypium from the genus Kokia and Gossypioides 5-10 million years ago (adapted from 7,51-54). Letters in parenthesis indicate the genome assignment for each species. Red text indicates cultivated species belonging to the genus Gossypium.Out of the more than 50 Gossypium species, only the allotetraploids G. hirsutum and G. barbadense, and the diploids G. arboreum and G. herbaceum are cultivated for their spinnable fibers. G. hirsutum, which is also known as Upland cotton, Long Staple cotton or Mexican cotton, occupy over 90% of the world cotton cultivation whereas G. barbadense, otherwise known as Sea Island cotton, Pima cotton or Egyptian cotton, contributes to 8% of the global cotton production. The cultivated diploid species provide approximately 2% of the world’s cotton and are cultivated in the more traditional growing areas of India, Pakistan, China, Bangladesh and Iran [24,51].
Based on genetic hybridization properties, Gossypium species are further grouped into the primary, secondary and tertiary gene pools. Both the cultivated (G. hirsutum and G. barbadense) and wild allotetraploids (G. tomentosum, G. mustelinum and G. darwinii) comprise the primary gene pool of cotton. The secondary gene pool includes the diploids having the A, B, D and F genomes, whereas the tertiary gene pool is composed of species with C, E, G and K genomes [7,55]. Genetic diversity studies using random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), simple sequence repeats (SSR) and/or single nucleotide polymorphisms (SNPs) indicate the availability of a tremendous amount of genetic variation among the different wild species, as well as among exotic subspecies of Gossypium [9,56-60]. This genetic diversity is reflected in the extensive variation in the gross morphology, maturity, photoperiodicity, yield potential, fiber quality, environmental adaptability and tolerance to pests and diseases that has been reported for the wild species of Gossypium (Table 1) [55,61]. To efficiently utilize this natural variation in cotton improvement, the genetic and molecular basis of phenotypic variations observed across the wild Gossypium germplasm need to be unlocked.
Based on genetic hybridization properties, Gossypium species are further grouped into the primary, secondary and tertiary gene pools. Both the cultivated (G. hirsutum and G. barbadense) and wild allotetraploids (G. tomentosum, G. mustelinum and G. darwinii) comprise the primary gene pool of cotton. The secondary gene pool includes the diploids having the A, B, D and F genomes, whereas the tertiary gene pool is composed of species with C, E, G and K genomes [7,55]. Genetic diversity studies using random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), simple sequence repeats (SSR) and/or single nucleotide polymorphisms (SNPs) indicate the availability of a tremendous amount of genetic variation among the different wild species, as well as among exotic subspecies of Gossypium [9,56-60]. This genetic diversity is reflected in the extensive variation in the gross morphology, maturity, photoperiodicity, yield potential, fiber quality, environmental adaptability and tolerance to pests and diseases that has been reported for the wild species of Gossypium (Table 1) [55,61]. To efficiently utilize this natural variation in cotton improvement, the genetic and molecular basis of phenotypic variations observed across the wild Gossypium germplasm need to be unlocked.
DEVELOPMENT OF EXOTIC GENETIC RESOURCES TOWARDS THE UTILIZATION OF WILD COTTON RELATIVES IN TRAIT IMPROVEMENT
The wild cotton germplasm has been recognized as a rich reservoir of genes underlying traits of agronomic importance and to some extent, has been tapped to improve the productivity and fiber quality of cultivated cotton. Wide hybridizations using exotic subspecies of G. hirsutum has successfully transferred Verticillium wilt resistance and salinity tolerance from G. hirsutum subsp. mexicanum var. nervosum to elite upland cotton cultivars [62]. Similarly, quantitative trait loci (QTLs) controlling fiber quality and yield potential have been identified from interspecific hybrids developed between G. hirsutumand the wild allotetraploids G. darwinii, G. mustelinum and G. tomentosum [15,20,63]. The use of genetic bridges to facilitate crossing between the two tri-species hybrids G. hirsutum x G. longicalyx x G. armourianum and G. hirsutum x G. longicalyx x G. herbaceum has successfully introgressed the reniform nematode (Rotylenchus reniformisi) resistance from G. longicalyx to G. hirsutum [64].
Despite the successful transfer of useful genes from a few wild species and subspecies into the cultivated cotton by conventional means, the extent of wide hybridizations within species of Gossypium has been limited. Inter- specific hybridization between the cultivated G. barbadense and G. hirsutum via conventional crosses has so far been unsuccessful [65,66]. Failure to develop interspecific hybrids between these two species may be attributed to the numerous genomic incompatibilities that result in sterility, cytological abnormalities, distorted segregation, hybrid breakdown, limited recombination between homologous chromosomes, and linkage drag that transfers undesirable traits along with the genes of interest in the wide hybrids [67-69].
As an alternative to conventional gene introgression, exotic libraries that provide opportunities to break up disadvantageous associations between traits so that beneficial genes can be moved across Gossypiumspecies from different gene pools have been generated. Chromosome segment substitution lines (CSSLs), monosomic alien addition lines (MAALs) and multi-parent advanced generation inter-crosses (MAGIC) are only few of the powerful genetic tools that can be used to identify and quantify the effects of specific alleles from wild relatives.
CSSLs are developed by hybridizations between an elite or adapted crop cultivar with a wild donor parent. Chromosome introgressions from the wild donor to the cultivated parent genome are commonly monitored using molecular markers. Each CSSL is selected to carry only a single chromosome introgression in a known locus within the genome. The whole genome of a wild donor parent is typically represented in a set of CSSLs composed of several lines [70]. The uniform genetic background of CSSLs provides the advantage of easily associating a phenotype with the introgressed chromosome segment, as well as identifying genes/QTLs using only simple statistical analysis [71].
In cotton, CSSL sets representing the whole genome of G. barbadense in the background of G. hirsutumhave been developed by various groups of researchers and used to have been used to identify and map QTLs controlling fiber yield and quality [72-76]. To date however, no other Gossypium species has been used as a donor in the development of CSSLs.
MAALs are also important genetic stocks that are derived from crosses between a crop and its wild ancestor. Development of MAALs usually requires embryo rescue of the interspecific hybrid before it aborts. A distinct characteristic of MAALs is the presence of a single chromosome from the wild ancestor in addition to the normal chromosome complement of a given crop. MAALs not only provide a convenient way of dissecting wild genomes into individual chromosome in a functional genomics background but also serve as bridges to transfer favorable genes from the wild to the cultivated species [77].
Morphological, cytological and molecular analysis using microsatellite markers have aided in the development of MAALs for G. anomalum, G. australe, G. sturtianum and G. somalense in the background of the upland cotton. G. hirsutum ,G. australe and G. somalense have been reported to possess tolerance to drought, whereas all four wild species have been documented to have resistance to a range of pests (reniform nematode, Egyptian and pink bollworm, springtails, aphids and/or mites) and diseases (Fusarium, Verticillium, cotton wilt and/or angular leaf spot) [28,46,50,78,79]. Although these MAALs have already been characterized morphologically and to some extent, physiologically, screening of these exotic libraries under a range of biotic and abiotic pressures would allow the identification of lines that can be used for the improvement of disease resistance and drought tolerance in cotton.
In contrast to CSSLs and MAALs which are based on bi-parental crosses, MAGIC populations involve intercrossing a number of parental lines for several generations to combine the genomes of all parents in the progeny lines [80]. The use of multiple parents to develop the population effectively increases the genetic variation within the population as a result of greater mixing of diverse alleles. Because the population undergoes a greater number of recombination events, MAGIC populations can provide higher resolution for QTL mapping [81].
MAGIC populations have also been developed in cotton by intercrossing 10 cultivars and one non-commercial variety of G. hirsutum in a half-diallel design. Molecular characterization of the MAGIC population using SSRs and SNP markers showed introgressions coming from the 11 parents used in the crosses. Genome-wide association studies using this MAGIC population identified a QTL cluster controlling four fiber quality traits [82,83].
Despite the potential of MAGIC populations in moving genes from multiple wild cotton species into progeny lines, this type of genetic resource in cotton is still in its infancy. The value of utilizing wild MAGIC populations in the elucidation of genetic determinants underlying complex traits and in delivering solutions to current challenges in crop production is yet to be realized [80]. The potential use of wild MAGIC populations in actual breeding programs would also depend on the successful evaluation of the genetic diversity at the DNA level and linking these variations with observable phenotypic performances.
Aside from developing exotic libraries, biotechnological methods such as haploid induction, interspecific cell fusion and somatic hybridization have been proven effective in overcoming sexual incompatibilities between the wild and the cultivated cotton [42,84]. Symmetric electrofusions between the tetraploid G. hirsutum and the diploids G. trilobum, G. klotzschianum, G. bickii, G. davidsonii and G. stocksii have successfully generated viable somatic hybrids containing novel genetic combinations coming from the allotetraploid and the diploid parents [42,85-89]. The results of such studies indicate the feasibility and the efficiency of somatic hybridization in incorporating divergent genomes into breeding programs for cotton [42,44,45].
Despite the successful transfer of useful genes from a few wild species and subspecies into the cultivated cotton by conventional means, the extent of wide hybridizations within species of Gossypium has been limited. Inter- specific hybridization between the cultivated G. barbadense and G. hirsutum via conventional crosses has so far been unsuccessful [65,66]. Failure to develop interspecific hybrids between these two species may be attributed to the numerous genomic incompatibilities that result in sterility, cytological abnormalities, distorted segregation, hybrid breakdown, limited recombination between homologous chromosomes, and linkage drag that transfers undesirable traits along with the genes of interest in the wide hybrids [67-69].
As an alternative to conventional gene introgression, exotic libraries that provide opportunities to break up disadvantageous associations between traits so that beneficial genes can be moved across Gossypiumspecies from different gene pools have been generated. Chromosome segment substitution lines (CSSLs), monosomic alien addition lines (MAALs) and multi-parent advanced generation inter-crosses (MAGIC) are only few of the powerful genetic tools that can be used to identify and quantify the effects of specific alleles from wild relatives.
CSSLs are developed by hybridizations between an elite or adapted crop cultivar with a wild donor parent. Chromosome introgressions from the wild donor to the cultivated parent genome are commonly monitored using molecular markers. Each CSSL is selected to carry only a single chromosome introgression in a known locus within the genome. The whole genome of a wild donor parent is typically represented in a set of CSSLs composed of several lines [70]. The uniform genetic background of CSSLs provides the advantage of easily associating a phenotype with the introgressed chromosome segment, as well as identifying genes/QTLs using only simple statistical analysis [71].
In cotton, CSSL sets representing the whole genome of G. barbadense in the background of G. hirsutumhave been developed by various groups of researchers and used to have been used to identify and map QTLs controlling fiber yield and quality [72-76]. To date however, no other Gossypium species has been used as a donor in the development of CSSLs.
MAALs are also important genetic stocks that are derived from crosses between a crop and its wild ancestor. Development of MAALs usually requires embryo rescue of the interspecific hybrid before it aborts. A distinct characteristic of MAALs is the presence of a single chromosome from the wild ancestor in addition to the normal chromosome complement of a given crop. MAALs not only provide a convenient way of dissecting wild genomes into individual chromosome in a functional genomics background but also serve as bridges to transfer favorable genes from the wild to the cultivated species [77].
Morphological, cytological and molecular analysis using microsatellite markers have aided in the development of MAALs for G. anomalum, G. australe, G. sturtianum and G. somalense in the background of the upland cotton. G. hirsutum ,G. australe and G. somalense have been reported to possess tolerance to drought, whereas all four wild species have been documented to have resistance to a range of pests (reniform nematode, Egyptian and pink bollworm, springtails, aphids and/or mites) and diseases (Fusarium, Verticillium, cotton wilt and/or angular leaf spot) [28,46,50,78,79]. Although these MAALs have already been characterized morphologically and to some extent, physiologically, screening of these exotic libraries under a range of biotic and abiotic pressures would allow the identification of lines that can be used for the improvement of disease resistance and drought tolerance in cotton.
In contrast to CSSLs and MAALs which are based on bi-parental crosses, MAGIC populations involve intercrossing a number of parental lines for several generations to combine the genomes of all parents in the progeny lines [80]. The use of multiple parents to develop the population effectively increases the genetic variation within the population as a result of greater mixing of diverse alleles. Because the population undergoes a greater number of recombination events, MAGIC populations can provide higher resolution for QTL mapping [81].
MAGIC populations have also been developed in cotton by intercrossing 10 cultivars and one non-commercial variety of G. hirsutum in a half-diallel design. Molecular characterization of the MAGIC population using SSRs and SNP markers showed introgressions coming from the 11 parents used in the crosses. Genome-wide association studies using this MAGIC population identified a QTL cluster controlling four fiber quality traits [82,83].
Despite the potential of MAGIC populations in moving genes from multiple wild cotton species into progeny lines, this type of genetic resource in cotton is still in its infancy. The value of utilizing wild MAGIC populations in the elucidation of genetic determinants underlying complex traits and in delivering solutions to current challenges in crop production is yet to be realized [80]. The potential use of wild MAGIC populations in actual breeding programs would also depend on the successful evaluation of the genetic diversity at the DNA level and linking these variations with observable phenotypic performances.
Aside from developing exotic libraries, biotechnological methods such as haploid induction, interspecific cell fusion and somatic hybridization have been proven effective in overcoming sexual incompatibilities between the wild and the cultivated cotton [42,84]. Symmetric electrofusions between the tetraploid G. hirsutum and the diploids G. trilobum, G. klotzschianum, G. bickii, G. davidsonii and G. stocksii have successfully generated viable somatic hybrids containing novel genetic combinations coming from the allotetraploid and the diploid parents [42,85-89]. The results of such studies indicate the feasibility and the efficiency of somatic hybridization in incorporating divergent genomes into breeding programs for cotton [42,44,45].
UNDERSTANDING THE GENETIC BASIS OF DISEASE RESISTANCE AND SALT TOLERANCE IN WILD GOSSYPIUM SPECIES TOWARDS THEIR UTILIZATION IN BREEDING
Verticillium wilt resistance
Verticillium wilt is an important disease in cotton caused by the soil-borne fungus Verticillium dahliae. One of the salient features of this fungus is its ability to remain viable in the soil for extended periods, allowing the build-up of pathogen inoculums over time. Successful colonization of the host cotton plants by the fungal pathogen leads to systemic infection which can eventually result in leaf wilting, chlorosis and necrosis. Infection by the pathogen during the later stages of plant growth development can lead to floral organ abscission [90-92].
Cultural management, as well as application of fungicides and other soil amendments has been widely used to control Verticillium wilt in cotton. To date however, breeding and cultivation of resistant cultivars remain the most efficient, cost-effective and environmentally benign means to manage the disease [91].
G. barbadense has been reported to have higher levels of resistance to Verticillium wilt compared to the upland cotton G. hirsutum. Backcross inbred lines developed between the two allotetraploids successfully introgressed disease resistance mechanisms from Pima to the upland cotton, along with improved fiber quality. Commercial cotton cultivars developed by major seed companies in the US have also been reported to benefit from Verticillium wilt resistance from G. barbadense [16]. The widespread deployment of cultivars with Verticillium wilt resistance that is based only on the genetic variation provided by Pima cotton risks the breakdown of such resistance mechanism with the emergence of a new or more virulent pathotypes or races of the pathogens. For a broader spectrum and more durable form of resistance to Verticillium wilt, the wild G. thurberi, G. trilobum, and G. capitis-viridis can be tapped for novel genetic variation conferring resistance to the disease.
The wild species, G. thurberi (DD genome) has been identified to harbor genes conferring resistance to Verticillium wilt [28,33]. Proteomic studies on G. thurberi plants infected with V. dahlia identified 52 up-regulated proteins that are involved in stress and disease resistance, transcriptional regulation, signal transduction, protein processing and degradation, photosynthesis, production capacity and basic metabolism. While the result of the study indicates that disease response in G. thurberi is a function of the expression of multiple genes, it is important to note that five of the up-regulated proteins are products of disease resistance genes [33]. A reverse-genetics approach to identify the genes coding for the upregulated proteins will provide basis for further investigation on the mechanisms underlying Verticillium wilt resistance in G. thurberi, towards practical applications in cotton improvement.
Like G. thurberi, G. trilobum (DD genome) and G. capitis-viridis (BB genome) have been reported to have resistance to Verticillium wilt [30,42]. Initial efforts to utilize the important agronomic traits of G. trilobum and G. capitis-viridis in breeding for G. hirsutum improvement have not been successful due to reproductive barriers between the two wild species and the upland cotton. To circumvent difficulties in producing wide hybrids due to fertility barriers, protoplast fusion was used to obtain symmetric somatic hybrids between G. trilobum and G. hirsutum. Flow cytometry in combination with the use of molecular markers such as RAPDs, SSRs, AFLPs and sequence-related amplified polymorphic markers confirmed the somatic hybridity of the regenerated plants. The hexaploid fusion plants exhibited morphological stability throughout generations, as well as strong vegetative growth and photosynthetic capacity [42]. Although the somatic hybrids have yet to be screened for Verticillium wilt resistance, the success by which protoplast fusion was able to incorporate the genetic material of G. trilobum together with that of G. hirsutum opens up possibilities of introducing disease resistance from wild Gossypium species to elite cotton cultivars.
To tap into the genetic variation present in G. capitis-viridis, tri-species hybridization followed by chromosome doubling was used. The tetraploid G. hirsutum cv. TM1 was initially crossed with G. australe(GG genome), creating a triploid F1 hybrid which was then treated with colchicine to generate an allohexaploid. Crossing the allohexaploid with G. capitis-viridis using the pseudophyletic introgression method generated an allopolyploid which has genetic contributions from the three Gossypium species. Field evaluations showed that the trihybrid has an intermediate morphology compared to the three wild species used to generate the hybrid, as well as higher resistance to insect pests [30]. Screening of the tri-hybrid against Verticillium wilt will determine the utilization of this novel germplasm in breeding programs to improve the resistance of cotton to the disease.
Cultural management, as well as application of fungicides and other soil amendments has been widely used to control Verticillium wilt in cotton. To date however, breeding and cultivation of resistant cultivars remain the most efficient, cost-effective and environmentally benign means to manage the disease [91].
G. barbadense has been reported to have higher levels of resistance to Verticillium wilt compared to the upland cotton G. hirsutum. Backcross inbred lines developed between the two allotetraploids successfully introgressed disease resistance mechanisms from Pima to the upland cotton, along with improved fiber quality. Commercial cotton cultivars developed by major seed companies in the US have also been reported to benefit from Verticillium wilt resistance from G. barbadense [16]. The widespread deployment of cultivars with Verticillium wilt resistance that is based only on the genetic variation provided by Pima cotton risks the breakdown of such resistance mechanism with the emergence of a new or more virulent pathotypes or races of the pathogens. For a broader spectrum and more durable form of resistance to Verticillium wilt, the wild G. thurberi, G. trilobum, and G. capitis-viridis can be tapped for novel genetic variation conferring resistance to the disease.
The wild species, G. thurberi (DD genome) has been identified to harbor genes conferring resistance to Verticillium wilt [28,33]. Proteomic studies on G. thurberi plants infected with V. dahlia identified 52 up-regulated proteins that are involved in stress and disease resistance, transcriptional regulation, signal transduction, protein processing and degradation, photosynthesis, production capacity and basic metabolism. While the result of the study indicates that disease response in G. thurberi is a function of the expression of multiple genes, it is important to note that five of the up-regulated proteins are products of disease resistance genes [33]. A reverse-genetics approach to identify the genes coding for the upregulated proteins will provide basis for further investigation on the mechanisms underlying Verticillium wilt resistance in G. thurberi, towards practical applications in cotton improvement.
Like G. thurberi, G. trilobum (DD genome) and G. capitis-viridis (BB genome) have been reported to have resistance to Verticillium wilt [30,42]. Initial efforts to utilize the important agronomic traits of G. trilobum and G. capitis-viridis in breeding for G. hirsutum improvement have not been successful due to reproductive barriers between the two wild species and the upland cotton. To circumvent difficulties in producing wide hybrids due to fertility barriers, protoplast fusion was used to obtain symmetric somatic hybrids between G. trilobum and G. hirsutum. Flow cytometry in combination with the use of molecular markers such as RAPDs, SSRs, AFLPs and sequence-related amplified polymorphic markers confirmed the somatic hybridity of the regenerated plants. The hexaploid fusion plants exhibited morphological stability throughout generations, as well as strong vegetative growth and photosynthetic capacity [42]. Although the somatic hybrids have yet to be screened for Verticillium wilt resistance, the success by which protoplast fusion was able to incorporate the genetic material of G. trilobum together with that of G. hirsutum opens up possibilities of introducing disease resistance from wild Gossypium species to elite cotton cultivars.
To tap into the genetic variation present in G. capitis-viridis, tri-species hybridization followed by chromosome doubling was used. The tetraploid G. hirsutum cv. TM1 was initially crossed with G. australe(GG genome), creating a triploid F1 hybrid which was then treated with colchicine to generate an allohexaploid. Crossing the allohexaploid with G. capitis-viridis using the pseudophyletic introgression method generated an allopolyploid which has genetic contributions from the three Gossypium species. Field evaluations showed that the trihybrid has an intermediate morphology compared to the three wild species used to generate the hybrid, as well as higher resistance to insect pests [30]. Screening of the tri-hybrid against Verticillium wilt will determine the utilization of this novel germplasm in breeding programs to improve the resistance of cotton to the disease.
Salinity tolerance
The wild species of Gossypium serve as a treasure trove of genetic variation not only for resistance to pests and diseases but also for tolerance to a range of abiotic stresses including mild frost, drought and salinity. Due to the reproductive barriers that impedes the interspecific hybridization among genetically distant species within the genus Gossypium, wild cotton relatives have not been largely used in breeding programs. Despite this constraint, studies have been conducted to examine the complexity of salt tolerance mechanisms in wild cotton using various ‘omics’ strategies. Understanding the genetic and molecular mechanisms that regulate salt tolerance in the wild cotton species will provide the necessary first step in exploiting natural variation within Gossypium in breeding for salinity tolerance in cotton.
The wild species G. davidsonii (DD genome), G. klotzschianum (DD genome) and G. aridum (DD genome) have been reported to exhibit tolerance to salinity stress. Next-generation RNA-sequencing in G. davidsonii from Baja, California identified 4744 and 5337 differentially expressed genes in the roots and leaves, respectively, of plants subjected to salt stress. Functional annotation of these genes elucidated the role of the Salt Overly Sensitive (SOS) and Reactive Oxygen Species (ROS) signaling pathways in salt tolerance in G. davidsonii [36]. In G. klotzschianum plants subjected to high salt stress (300 mM NaCl), transcriptome analysis showed the differential expression of 8312 and 6732 genes in the roots and leaves, respectively. Similar to G. davidsonii, functional annotation of genes that were differentially expressed in G. klotzschianum under high salt stress were involved in ROS and SOS signaling pathways, with the addition of genes involved in hormone biosynthesis and signal transduction [37]. In both wild species, differential expression was also observed in the transcription factor families of AP2/EREBP, bZIP, bHLH, MYB, NAC, and WRKY . Transcriptome analysis in G. aridum plants subjected to salt stress (200 mM NaCl) detected differential expression of genes involved in transport, hormone-stimulus response and signaling [38].
Although the molecular basis of salt tolerance in wild cotton relatives are far from being understood, results of ‘omics’ studies to dissect the complexity of salt tolerance in these wild species will provide basis for further investigations towards the identification and validation of genes/quantitative trait loci conferring salt tolerance in cotton.
The wild species G. davidsonii (DD genome), G. klotzschianum (DD genome) and G. aridum (DD genome) have been reported to exhibit tolerance to salinity stress. Next-generation RNA-sequencing in G. davidsonii from Baja, California identified 4744 and 5337 differentially expressed genes in the roots and leaves, respectively, of plants subjected to salt stress. Functional annotation of these genes elucidated the role of the Salt Overly Sensitive (SOS) and Reactive Oxygen Species (ROS) signaling pathways in salt tolerance in G. davidsonii [36]. In G. klotzschianum plants subjected to high salt stress (300 mM NaCl), transcriptome analysis showed the differential expression of 8312 and 6732 genes in the roots and leaves, respectively. Similar to G. davidsonii, functional annotation of genes that were differentially expressed in G. klotzschianum under high salt stress were involved in ROS and SOS signaling pathways, with the addition of genes involved in hormone biosynthesis and signal transduction [37]. In both wild species, differential expression was also observed in the transcription factor families of AP2/EREBP, bZIP, bHLH, MYB, NAC, and WRKY . Transcriptome analysis in G. aridum plants subjected to salt stress (200 mM NaCl) detected differential expression of genes involved in transport, hormone-stimulus response and signaling [38].
Although the molecular basis of salt tolerance in wild cotton relatives are far from being understood, results of ‘omics’ studies to dissect the complexity of salt tolerance in these wild species will provide basis for further investigations towards the identification and validation of genes/quantitative trait loci conferring salt tolerance in cotton.
Conclusion
Crop failures due to generations of utilizing a narrow genetic base for various crops have been documented throughout the history of agriculture. The wild species of Gossypium offers a tremendous amount of genetic variation that can be exploited to create and select for novel gene combinations that can give the highest productivity under a wide range of agricultural environments. The initial hurdle in developing interspecific cotton hybrids due to cross incompatibilities has been overcome using a suite of conventional and biotechnological tools that facilitated the effective incorporation of genetic materials from the wild Gossypium germplasm into existing cotton cultivars. Despite this major accomplishment, a large portion of the natural variation in the wild species of cotton remains unexploited. A requisite in the effective utilization of the rich diversity present in the wild cotton germplasm is the elucidation of the genetic basis of important agronomic traits. Recent advances in genomic technologies including the advent of high throughput sequencing technologies and genotyping platforms will allow rapid gene discovery towards a more precise utilization of natural genetic variation in cotton improvement.
REFERENCES
- Spindel J, McCouch S (2017) Ensuring and exploiting genetic diversity in rice. In: Sasaki T (ed.). Achieving sustainable cultivation of rice, (Vol 1). Breeding for higher yield and quality. Taylor & Francis Group, London, UK.
- Hwang YT, Wijekoon C, Kalischuk M, Johnson D, Howard R, et al. (2014) Evolution and management of Irish potato famine pathogen Phytophthora infestans in Canada and the United States. Am J Potato Res 91: 579-593.
- Ordonez N, Seidl MF, Waalwijk C, Drenth A, Kilian A, et al. (2015) Worse comes to worst: Bananas and panama disease - When plant and pathogen clones meet. PLoS Pathog 11: 1005197.
- Bruns HA (2017) Southern corn leaf blight: A story worth telling. Agro J 109: 1-7.
- Agrios GN (2005) Plant diseases caused by fungi. In: Agrios GN (ed.). Plant Pathology, (5th edn). Elsevier, New York, USA.
- Granett J, Walker A, De Benedictis J, Fong G, Lin H, et al. (1996) California grape phylloxera more viable than expected. California Agri 50: 9-13.
- Campbell BT, Saha S, Percy R, Frelichowski J, Jenkind JN, et al. (2010) Status of the global cotton germplasm resource. Crop Sci 50: 1161-1179.
- http://www.ers.usda.gov/data-products/adoption-of-genetically-engineered-crops-in-the-us/recent-trends-in-ge-adoption.aspx.
- Hinze LL, Hulse-Kemp AM, Wilson IW, Zhu QH, Llewellyn DJ, et al. (2017) Diversity analayis of cotton (Gossypium hirsutum L.) germplasm using the CottonSNP63K array. BMC Plant Biol 17: 37.
- Alonso-Blanco C, Mendez-Vigo B, Koornneef M (2005) From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. Int J Dev Biol 49: 717-732.
- Redden R (2013) New approaches for crop genetic adaptation to the abiotic stresses predicted with climate change. Agron 3: 419-432.
- Rick CM (1988) Tomato-like nightshade: Affinities, auto-ecology and breeders’ opportunities. Econ Bot 42: 145-154.
- Canady MA, Meglic V, Chetalat RT (2005) A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome 48: 685-697.
- Nevo E, Chen G (2010) Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ 33: 670-685.
- Keerio AA, Shen C, Nie Y, Ahmed MM, Zhang X, et al. (2018) QTL mapping of fiber quality and yield traits based on introgression lines derived from Gossypium hirsutum × G. tomentosum. Int J Mol Sci 19: 243.
- Zhang J, Sanogo S, Flynn R, Baral J, Bajaj S, et al. (2012) Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 187: 147-160.
- Saha S, Raska DA, Stelly DM (2006) Upland cotton (Gossypium hirsutum L.) × hawaiian cotton (G. tomentosum Nutt. Ex Seem.) F1 hybrid hypoaneuploid chromosome substitution series. J of Cotton Sci 10: 263-272.
- Hulse-Kemp AM, Ashrafi H, Zheng X, Wang F, Hoegenauer KA, et al. (2014) Development and bin mapping of gene-associated interspecific SNPs for cotton (Gossypium hirsutum L.) introgression breeding efforts. BMC Genomics 15: 945.
- Zhang Z, Rong J, Waghmare VN, Chee PW, May OL, et al. (2011) QTL alleles for improved fiber quality from a wild hawaiian cotton, Gossypium tomentosum. Theor Appl Genet 123: 1075-1088.
- Wang B, Liu L, Zhang D, Zhuang Z, Guo H, et al. (2016) A genetic map between Gossypium hirsutum and the Brazilian endemic G. mustelinum and its application to QTL mapping. G3 (Bethesda) 6: 1673-1685.
- Wendel JF, Rowley R, Stewart JMD (1994) Genetic diversity in and phylogenetic relationships of the Brazilian endemic cotton, Gossypium mustelinum (Malvaceae). Pl Syst Evol 192: 49-59.
- Chen H, Khan MK, Zhou Z, Wang X, Cai X, et al. (2015) A high-density SSR genetic map constructed from a F2 population of Gossypium hirsutum and Gossypium darwinii. Gene 574: 273-286.
- Chiavegato EJ, Gridi-Papp IL, Kondo JI, Medina DM (1985) Interespecific hybridization between Gossypium hirsutum var. latifolium and G. herbaceum var. africanum. Bragantia 44: 629-643.
- Kulkarni VN, Khadi BM, Maralappanavar MS, Deshapande LA, Narayanan SS (2009) The worldwide gene pools of Gossypium arboreum L. and G. herbaceum L. and their improvement In: Paterson AH (ed.). Genetics and and Genomics of Cotton, (Vol 3). Springer Science & Business Media, New York, USA.
- Stanton M, Tugwell NP, McD Stewart J (1992) Evaluation of Gossypium arboreum L. germplasm for resistance to thrips. Genet Resour and Crop Evol 39: 89-95.
- Kantartzi SK, Ulloa M, Sacks E, Stewart JM (2009) Assessing genetic diversity in Gossypiumarboreum L. cultivars using genomic and EST-derived microsatellites. Genetica 136: 141-147.
- Miyazaki J, Stiller WN, Wilson LJ (2012) Novel cotton germplasm with host plant resistance to twospotted spider mite. Field Crops Res 134: 114-121.
- Wang X, Wang Y, Wang C, Chen Y, Chen Y, et al. (2016) Characterization of eleven monosomic alien addition lines added from Gossypium anomalum to Gossypium hirsutum using improved GISH and SSR markers. BMC Plant Biol 16: 218.
- Wendel JF, Grover CE (2015) Taxonomy and evolution of the cotton genus, Gossypium. In: Fang DD, Percy RG (eds.). Cotton, (2nd edn). Am Soc Agron, Wisconsin, USA.
- Chen D, Wu Y, Zhang X, Li F (2015) Analysis of [Gossypium capitis-viridis × (G. hirsutum × G.australe)2] trispecific hybrid and selected characteristics. PloS One 10: 0127023.
- Bi IV, Baudoin JP, Mergeai G (1998) Cytogenetics of the ‘glandless-seed and glanded-plant’ trait from Gossypium sturtianum willis introgressed into upland cotton (Gossypium hirsutum L.). Plant Breed 117: 235-241.
- Becerra Lopez-Lavalle LA, McFadden H, Brubaker CL (2007) The effect of Gossypium C-genome chromosomes on resistance to Fusarium wilt in allotetraploid cotton. Theor Appl Genet 115: 477-488.
- Zhao F, Fang W, Xie E, Zhao Y, Tang Z, et al. (2012) Proteomic identification of differentially expressed proteins in Gossypium thurberi inoculated with cotton Verticillium dahliae. Plant Sci 185-186:176-184.
- Pushpam R, Raveendran TS (2006) Production of interspecific hybrids between Gossypium hirsutum and jassid resistant wild species G. raimondii and G. armourianum. Cytologia 71: 407-418.
- Meyer VG (1975) Male sterility from Gossypium harknessii. J Hered 66: 23-27.
- Zhang F, Zhu G, Du L, Shang X, Cheng C, et al. (2016) Genetics regulation of salt stress tolerance revealed by RNA-Seq in cotton diploid wild species, Gossypium davidsonii. Sci Rep 6: 20582.
- Wei Y, Xu Y, Lu P, Wang X, Li Z, et al. (2017) Salt stress responsiveness of a wild cotton species (Gossypium klotzschianum) based on transcriptomic analysis. PloS One 12: 0178313.
- Xu P, Liu Z, Fan X, Gao J, Zhang X, et al. (2013) de novo transcriptome sequencing and comparative analysis of differentially expressed genes in Gossypium aridum under salt stress. Gene 525: 26-34.
- Sacks EJ, Robinson AF (2009) Introgression of resistance to reniform nematode (Rotylenchulus reniformis) into upland cotton (Gossypium hirsutum) from Gossypium arboreum and a G. hirsutum/Gossypium aridum bridging line. Field Crops Res 112: 1-6.
- Azhar M, Anjum Z, Mansoor S (2013) Gossypium gossypioides: A source of resistance against cotton leaf curl disease among D genome diploid cotton species. J of Anim Plant Sci 23: 1436-1440.
- Peggy GF, Brady BL (2002) Verticillium wilts. CABI 10: 256.
- Yu XS, Chu BJ, Liu RE, Sun LJ, Brian JJ, et al. (2012) Characteristics of fertile somatic hybrids of G. hirsutum L. and G. trilobum generated via protoplast fusion. Theor Appl Genet 125: 1503-1516.
- Chen Y, Chen Y, Feng S, Zhao T, Zhou B (2018) Overcoming obstacles to interspecific hybridization between Gossypium hirsutum and G. turneri. Euphytica 214: 35.
- Nazeer W, Ahmad S, Mahmood K, Tipu AL, Mahmood A, et al. (2014) Introgression of genes for cotton leaf curl virus resistance and increased fiber strength from Gossypium stocksii into upland cotton (G. hirsutum). Genet Mol Res 13: 1133-1143.
- Yik CP, Birchfield W (1984) Resistant germplasm in Gossypium species and related plants to Rotylenchulus reniformis. J Nematol 16: 146-153.
- Zhou Z, Yu P, Liu G, He J, Chen J, et al. (2004) Morphological and molecular characterization of two G. somalense Monosomic Alien Addition Lines (MAALs). Chin Sci Bull 49: 910-914.
- Dighe ND, Robinson AF, Bell AA, Menz MA, Cantrell RG, et al. (2009) Linkage mapping of resistance to reniform nematode in cotton following introgression from Gossypium longicalyx (Hutch. and Lee). Crop Sci 49: 1151-1164.
- Zhu S, Jiang Y, Reddy N (2005) Introgression of a gene for delayed pigment gland morphogenesis from Gossypium bickii into upland cotton. Plant Breed 124: 590-594.
- Schuster M, Jenkins J, Maxwell F (1972) Resistance to the two spotted spider mite in certain Gossypium hirsutum races, Gossypium species, and glanded-glandless counterpart cottons. J Econ Entomol 65: 1108-1110.
- Chen Y, Wang Y, Wang K, Zhu X, Guo W, et al. (2014) Construction of a complete set of alien chromosome addition lines from Gossypium australe in Gossypium hirsutum: Morphological, cytological, and genotypic characterization. Theor Appl Genet 127: 1105-1121.
- Wendel JF, Brubaker CL, Seelanan T (2010) The origin and evolution of Gossypium. In: Stewart, JM, Oosterhius D, Heitholt JJ, Mauney JR (eds.). Physiology of cotton. Springer, Dordrecht, Netherlands.
- Phillips LL (1964) Segregation in new allopolyploids of Gossypium. V. Multivalent formation in New World × Asiatic and New World × wild American hexaploid. Am J Bot 51: 324-329.
- Zhang HB, Li Y, Wang B, Chee PW (2008) Recent advances in cotton genomics. Int J Plant Genomics 2008: 724304.
- Gallagher JP, Grover CE, Rex K, Moran M, Wendel JF (2017) A new species of cotton from Wake Atoll, Gossypium stephensii (Malvaceae). Syst Bot 42: 115-123.
- Abdurakhmonov IY, Buriev ZT, Shermatov SE, Abdullaev AA, Urmonov K, et al. (2012) Genetic diversity in Gossypium genus. In: Caliksan M (ed.). Genetic diversity in plants. InTech, Uzbekistan, Central Asia.
- Tiwari RS, Stewart JMcD (2008) Molecular diversity and determination of hybridization among the G-genome Gossypium species. In: Oosterhuis DM (ed.). Summaries of Arkansas cotton research, Research series 573.
- Barroso PAV, Hoffman LV, de Freitas RB, de Batista ACE, Alves MF, et al. (2010) In situ conservation and genetic diversity of three populations of Gossypium mustelinum Miers ex Watt. Genet Resour Crop Evol 57: 343-349.
- Zhang Y, Wang XF, Li ZK, Zhang GY, Ma ZY (2011) Assessing genetic diversity of cotton cultivars using genomic and newly developed expressed sequence tag-derived microsatellite markers. Genet Mol Res 10: 1462-1470.
- Sethi K, Siwach P, Verma SK (2015) Assessing genetic diversity among six populations of Gossypiumarboreum L. using mircrosatellite markers. Physiol Mol Biol Plants 21: 531-539.
- Hinze LL, Gazave E, Gore MA, Fang DD, Scheffler BE, et al. (2016) Genetic diversity of the two commercial tetraploid cotton species in the Gossypium diversity reference set. J Hered 107: 274-286.
- Lee JA, Fang DD (2015) Cotton as a world crop: Origin, history and current status. In: Fang DD, Percy RG (eds). Cotton, (2nd edn). American Society of Agronomy, Wisconsin, USA.
- Abdurakhmonov IY (2007) Exploiting genetic diversity. Proceedings of World Cotton Research Conference-4, P2153, Lubbock, Texas, USA 10-14.
- Wang B, Nie Y, Lin Z, Zhang X, Liu J, et al. (2012) Molecular diversity, genomic constitution, and QTL mapping of fiber quality by mapped SSRs in introgression lines derived from Gossypium hirsutum × G. darwinii Watt. Theor Appl Genet 125: 1263-1274.
- Robinson AF, Bell AA, Dighe ND, Menz MA, Nichols RL, et al. (2007) Introgression of resistance to nematode Rotylenchulus reniformis into upland cotton (Gossypium hirsutum) from Gossypiumlongicalyx. Crop Sci 47: 1865-1877.
- Lacape J, Nguyen T, Courtois B, Belot J, Giband M, et al. (2005) QTL analysis of cotton fiber quality using multiple Gossypium hirsutum × Gossypium barbadense backcross generations. Crop Sci 45: 123-140.
- He DH, Lin ZX, Zhang XL, Zhang YX, Li W, et al. (2008) Dissection of genetic variance of fiber quality in advanced generations from an interspecific cross of Gossypium hirsutum and G. barbadense. Plant Breed 127: 286-294.
- Reinisch AJ, Dong J, Brubaker CL, Stelly DM, Wendel JF, et al. (1994) A detailed RFLP map of cotton, Gossypium hirsutum × Gossypium barbadense: Chromosome organization. Genetics 138: 829-847.
- Ulloa M, Saha S, Jenkins JN, Meredith WR, McCarty JC, et al. (2005) Chromosomal assignment of RFLP linkage groups harboring important QTLs on an intraspecific cotton (Gossypium hirsutum) joinmap. J Hered 96: 132-144.
- Khush G, Brar DS (2017) Alien introgression in rice. Nucleus 60: 251-261.
- Ali ML, Sanchez PL, Yu S, Lorieux M, Eizenga GC (2010) Chromosome segment substitution lines: A powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 3: 218-234.
- Gutierrez AG, Carabali SJ, Giraldo OX, Martinez CP, Correa F, et al. (2010) Identification of a rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa × Oryza glaberrima introgression lines. BMC Plant Biol 10: 6.
- Zhai H, Gong W, Tan Y, Liu A, Song W, et al. (2016) Identification of chromosome segment substitution lines of Gossypium barbadense introgressed in G. hirsutum and quantitative trait locus mapping for fiber quality and yield traits. PLoS One 11: 0159101.
- Stelly DM, Saha S, Raska DA, Jenkins JN, McCarty JC, et al. (2005) Registration of 17 upland (Gossypium hirsutum) germ-plasm lines disomic for different G. barbadense chromosome orarm substitutions. Crop Sci 45: 2663-2665.
- Saha S, Wu J, Jenkins JN, McCarty JC, Hayes R, et al. (2010) Genetic dissection of chromosome substitution lines of cotton to discover novel Gossypium barbadense L. alleles for improvement of agronomic traits. Theor Appl Genet 120: 1193-1205.
- Saha S, Wu J, Jenkins JN, McCarty JC, Stelly DM (2013) Interspecific chromosomal effects on agronomic traits in Gossypium hirsutum by AD analysis using intermated G. barbadense chromosome substitution lines. Theor Appl Genet 126: 109-117.
- Li PT, Wang M, Lu QW, Ge Q, Rashid MHO, et al. (2017) Comparative transcriptome analysis of cotton fiber development of upland cotton Gossypium hirsutum) and chromosome segment substitution lines from G. hirsutum × G. barbadense. BMC Genomics 18: 705.
- Brar DS, Khush GS (1997) Alien introgression in rice. Plant Mol Biol 35: 35-47.
- Rooney WL, Stelly DM, Altman DW (1991) Identification of four Gossypium sturtianum monosomic alien addition derivatives from a backcrossing program with G. hirsutum. Crop Sci 31: 337-341.
- Sarr D, Lacape J, Jacquemin J, Benbouza H, Toussaint A, Baudoin J, et al. (2012) Alien chromosome transmission and somatic elimination in monosomic addition lines of Gossypium australe F. Muell in G. hirsutum L. Euphytica 183: 55-64.
- Huang BE, Verbyla KL, Verbyla AP, Raghavan C, Singh VK, et al. (2015) MAGIC populations in crops: Current status and future prospects. Theor Appl Genet 128: 999-1017.
- Huang BE, George AW, Forrest KL, Kilian A, Hayden AJ, et al. (2012) A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotech J 10: 826-839.
- Islam MS, Thyssen GN, Jenkins JN, Zeng L, Delhom CD, et al. (2016) A MAGIC population-based genome-wide association study reveals functional association of GhRBB1_A07 gene with superior fiber quality in cotton. BMC Genomics 17: 903.
- Li DG, Li ZX, Hu JS, Lin ZX, Li XF (2016) Polymorphism analysis of multi-parent advanced generation inter-cross (MAGIC) populations of upland cotton developed in China. Genet Mol Res 15.
- Multani DS, Khush GS, delos Reyes BG, Brar DS (2003) Alien genes introgression and development of monosomic alien addition lines from Oryza latifolia Desv. to rice, Oryza sativa L. Theor Appl Genet 107: 395-405.
- Sun YQ, Zhang XL, Nie YC, Guo XP, Jin SX, et al. (2004) Production and characterization of somatic hybrids between upland cotton (Gossypium hirsutum) and wild cotton (G. klotzschianum Anderss) via electrofusion. Theor Appl Genet 109: 472-479.
- Sun YQ, Zhang XL, Nie YC, Guo XP (2005) Production of fertile somatic hybrids of Gossypiumhirsutum + G. bickii and G. hirsutum + G. stockii via protoplast fusion. Plant Cell Tiss Organ Cult 83: 303-310.
- Sun YQ, Zhang XL (2006) Somatic hybrids between Gossypium hirsutum L. (4×) and G. davidsonii Kellog (2×) produced byprotoplast fusion. Euphytica 151: 393-400.
- Sun YQ, Liu SM, Wang Y, Brian JJ, Wang HZ, et al. (2011) An interspecific somatic hybrid between upland cotton (G. hirsutum L. cv. ZDM-3) and wild diploid cotton (G. klotzschianum A.). Plant Cell Tiss Organ Cult 106: 425-433.
- Yang XY, Zhang XL, Jin SX, Fu LL, Wang LG (2007) Production and characterization of asymmetric hybrids between upland cotton Coker 201 (Gossypium hirsutum) and wild cotton (G. klozschianum Anderss). Plant Cell Tiss Organ Cult 89: 225-235.
- Hampton RE, Wullschleger SD, Oosterhuis (1990) Impact of Verticillium wilt on net photosynthesis, respiration and photorespiration in field-grown cotton (Gossypium hirsutum L.). Physiol Mol Plant Pathol 37: 271-280.
- Zhou H, Fang H, Sanogo S, Hughs SE, Jones DC, et al. (2014) Evaluation of Verticillium wilt resistance in commercial cultivars and advanced breeding lines of cotton. Euphytica 196: 437-448.
- Mert M, Kurt S, Gencer O, Akiscan Y, Boyaci K, et al. (2005) Inheritance of resistance to Verticilliumwilt (Verticillium dahliae) in cotton (Gossypium hirsutum L.). Plant Breed 124: 102-104.
Citation: Shim J, Mangat PK, Angeles-Shim RB (2018) Natural variation in wild Gossypium species as a tool to broaden the genetic base of cultivated cotton. J Plant Sci Curr Res 2: 005.
Copyright: © 2018 Junghyun Shim, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

Journal Highlights
© 2026, Copyrights Herald Scholarly Open Access. All Rights Reserved!
