
OMICS Integration Studies for New Insights on Atopic Dermatitis
*Corresponding Author(s):
Yong-Doo ParkZhejiang Provincial Key Laboratory Of Applied Enzymology, Yangtze Delta Region Institute Of Tsinghua University, Jiaxing, China
Tel:+8657382582762,
Fax:+8657382582767
Email:parkyd@hotmail.com
Jun-Mo Yang
Department Of Dermatology, Samsung Medical Center, Sungkyunkwan University School Of Medicine, Seoul, Republic Of Korea
Tel:+82234103541,
Fax:+82234103869;
Email:junmo.yang@samsung.com
Abstract
The number of Atopic Dermatitis (AD) patients has increased with modernization and industrialization, and AD patients and their family members experience severe quality of life issues. It is generally recognized that a combination of environmental factors such as allergens and bacterial infections, as well as genetic factors, are the principal inducers of the multiple immunologic and inflammatory responses seen in AD patients. In the present review article, we focused on the following primary issues from the broad category of AD disease that have been reported in research articles, including those by the author's own research groups: i) AD types: characteristics of extrinsic and intrinsic types; ii) current treatments of AD and clinical applications; iii) new insights from OMICS integration studies for detecting hub genes; iv) summary of AD pathogenesis and associated factors; and v) further speculation for AD studies. The topics in this review that we focused on are expected to contribute to the understanding of AD disease-related factors. Since the mechanism of AD disease has been shown to be complex, our results may provide new clues to aid understanding of AD from the aspect of laboratorial OMICS applications.
Keywords
ABBREVIATIONS
AD: Atopic Dermatitis
HTS: High-Throughput Screening
PPI: Protein-Protein Interaction
PSIMAP: Protein Structural Interactome Map
PEIMAP: Protein Experimental Interactome Map
INTRODUCTION
There are more known diseases of the skin than any other organ system, as the skin is the first defense system against the external environment and frequently responds to various outer challenges. In this regard, studies on skin diseases-associated factors are important in order to understand the complex nature of defense mechanisms and various pathogeneses. The rapid development of OMICS, including proteome-based proteomics, genome-based genomics, and interactome-based interactomics, has enabled dermatologic researchers to understand skin diseases and diseases-associated factors [1,2]. The term 'OMICS' refers to the research areas ending in-omics such as proteomics, metabolomics, genomics, lipidomics, foodomics, and transcriptomics.
Recently, with the efforts of researchers and clinicians, several important Atopic Dermatitis (AD) diseases-associated factors have been identified; in particular, OMICS integration studies have successfully revealed new factors associated with the complexity of AD [3-5]. For dermatologic researchers of AD, new methods and technologies will be important to gain greater insight into the complex pathogenesis of AD and to develop effective and precise therapeutic strategies. The OMICS approach is an initial step used to identify candidate gene/protein pools of AD. Rapid confirmation by real-time PCR, RT-PCR, western blotting, and ELISA detection in patient-derived samples as a sequential study could validate observed OMICS data to confirm at an early stage the helpful methods to select candidates for biomarkers or target gene/proteins of interest. After determining the target proteins, functional studies can be conducted using cell or animal models, as well as large-scale studies on clinical samples from AD patients. Final confirmation and AD mechanisms may then be more clearly evaluated during the development of effective drugs to treat AD in clinical trials.
AD TYPES
Characteristics of extrinsic and intrinsic types
Current clinical treatments of AD
The above knowledge has been used to develop clinical treatments and associated medicines for treatment of AD patients [13-15]. These include 1) anti-IgE drugs for allergic responses such as omalizumab, humanized IgG1 monoclonal antibody; 2) emollient treatment for dry skin such as ceramide and its derivatives; 3) topical calcineurin inhibitors such as tacrolimus (FK506) and pimecrolimus for inhibitory effects on the activation of T cells and mast cells; 4) antipruritic agent, such as a combination of topical doxepin and topical corticosteroid for the relief of the itch-scratch cycle or capsaicin for reducing the itch sensation; 5) anti-inflammatory agents such as topical corticosteroids and TNF inhibitors; 6) antimicrobial antibiotics; and 7) UV light therapy. The AD patient population has greatly increased in almost every developed country regardless of the therapeutic efforts currently used to treat AD patients. Furthermore, more significant and effective treatments providing quick relief of severe symptoms such as itching still need to be developed. Moreover, specific diagnosis and case sensitive treatment are needed for different ages and types of AD patients. Thus, more insights are needed into the pathogenesis of AD, in addition to elucidating the immunologic aspects and genetic background of the disease. To achieve this goal, High-Throughput Screening (HTS)-based OMICS studies are required to obtain new information. The OMICS used for studying AD can be categorized by several developed methods such as proteomics, chip-based genomics such as Single-Nucleotide Polymorphism (SNP) studies, and interactomics of bioinformatics. Now, more than ever, the rapid development of HTS-based techniques has enabled researchers to understand AD disease-associated factors, regardless of their own drawbacks. New methods and technologies will be important to gain more insight into the complex pathogenesis of AD and to develop effective and individual therapeutic strategies. Still, it is generally recognized that HTS-based studies have their own drawbacks, which include discrepancy between translational and transcriptional levels of detected proteins, a relatively high cost for equipping and maintaining instruments, a prohibitively large size of candidate genes/proteins even after significant cut-off of data handling, and a limitation in applicable sample size. These problems can be improved by several solutions. For instance, sometimes minimizing patient-to-patient variation is used to try and overcome difficulties in the pooling method. The selection of interesting proteins/genes among a large sample size can be contextually consulted from previous reported publications. In addition, researchers could combine computational predictions via Protein-Protein Interaction (PPI) mapping of interactomics to help determine target proteins for subsequent studies. The adaptation of bioinformatic tools is a time- and cost-saving approach for the next step of functional and large-scale clinical studies. In this regards, an OMICS integration study based on the data found in HTS approaches could be extremely useful for the identification of AD questions for diagnostic and clinical purposes.
At the end of any AD research process or clinical trial, pruritus in AD patients should be considered the top priority. For this, pruritus-associated neurofibers and dry skin itching problems should receive more focus in further studies than any other AD factors.
NEW INSIGHTS FROM OMICS INTEGRATION STUDIES FOR DETECTING HUB (CORE) GENES
The HTS applications have the potential to produce many data pools that could make interactomic analyses. Among several methods, there are currently two kinds of algorithms such as the Protein Structural Interactome MAP (PSIMAP) and Protein Experimental Interactome MAP (PEIMAP) that have been applied to analyze HTS data pools (to see http://www.ncbi.nlm.nih.gov/pubmed/18215330). PSIMAP is a method that uses the structural domain of the Structural Classification of Proteins (SCOP) database; thus, the basic procedure of PSIMAP is to infer interactions between proteins by using their homologs and as such PSIMAP provides a structure-based interaction prediction. Meanwhile, PEIMAP is constructed by combining several experimental protein-protein interaction databases; thus, it is carried out as a redundancy check to remove identical protein sequences from the source interaction databases, including functionally classified protein-protein interactions. The PPIs can be used by researchers to determine protein targets for regulating AD-related pathways. These bioinformatic tools may help to identify core proteins among the detected candidate genes/proteins and explain their undetermined roles in AD. Therefore, several hub genes predicted from the network of AD-related factors can be useful as target genes for further study.
In the present review, we have successfully classified several important hub genes by comparing previously reported results of PPI mappings [16-20], and they are listed in Table 1. We found that SFN, YWHAE, C1R, TNC, BARD1, SEMA4F, and LPA have been selected as important hub genes since they were found multiple times using different methods and samples during AD research. The candidate proteins listed in Table 1 cited from previous results may provide important information to help elucidate the complex nature of AD disease. In respect to the comparison between transcriptional and translational levels of hub genes, the selected genes were well matched in both levels.
| Gene symbol | Gene name | Sample source | Detection Method | Cited references |
| SFN | Stratifin | Biopsy tissues | 34K-oligo gene chip/Affymetrix gene chip | [18,19] |
| YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | Biopsy tissues/Primary cultured fibroblasts | Affymetrix gene chip/Free-flow electrophoresis | [19,20] |
| C1R | Complement component 1, r subcomponent | Primary cultured fibroblasts | 8K-cDNA gene chip/Free-flow electrophoresis | [17,20] |
| TNC | Tenascin C | Primary cultured fibroblasts | 8K-cDNA gene chip/Free-flow electrophoresis | [17,20] |
| BARD1 | BRCA1 associated RING domain 1 | Primary cultured fibroblasts/Primary cultured keratinocytes | 8K-cDNA gene chip/Free-flow electrophoresis | [17,20] |
| SEMA4F | Semaphorin 4F | Primary cultured fibroblasts | 8K-cDNA gene chip/Free-flow electrophoresis | [17,20] |
| LPA | Apolipoprotein A | Primary cultured fibroblasts | 8K-cDNA gene chip/Free-flow electrophoresis | [17,20] |
Table 1: The hub genes/proteins predicted via PPI mappings from HTS results.
Stratifin (SFN) was detected earlier in ADE patient-derived skin tissues as a meaningful candidate gene but was not predicted as a hub gene. It was subsequently detected in AD patient-derived tissues as a hub gene in another DNA-chip-based microarray study. The real-time PCR analyses showed that the transcriptional level of SFN is conspicuously upregulated 4.5-fold in ADI patient-derived keratinocytes. In the epidermis, SFN is known to stimulate the syntheses of ECM factors (collagen, proteoglycans, and fibronectin) and the matrix metalloproteinases [21-23]. SFN is also known to be involved in the response to DNA damage, and this protein is involved in the cell cycle (G2 arrest). The lack of SFN expression due to hypermethylation has been reported in breast cancer. Thus, upregulation of SFN is an interesting target point to establish new therapeutic applications to control the process of wound healing for AD patients. The predicted SFN-associated PPI networks are shown in Figure 1.
Accordingly, it is also worthy to pay attention to YWHAE (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide), which was predicted as a hub gene along with SFN. YWHAE was detected in subsequent proteomic studies such as Free-Flow Electrophoresis (FFE) as an important hub protein. Both SFN and YWHAE were not well elucidated in AD pathogenesis until now and thus they could be potent target proteins among many candidate genes found in AD patient-derived samples. YWHAE has been shown to play important roles in cell division, signal transduction, and the pathogenesis of certain diseases; however, its role in AD has not yet been reported.
We were able to confirm that some hub genes were associated with AD pathogenesis that were previously reported, one of which had already been used as a biomarker for the diagnosis of AD disease. Such genes included FceRI (Fc fragment of IgE high affinity I receptor), which is shown in Figure 1A as a hub gene. FceRI is well-known as a factor that responds against external inducers such as allergens or bacterial infections that could trigger the pathogenesis of AD in a sequential way. Specifically dendritic cells take up allergens and induce T cell differentiation into Th2 cells. Th2 cells further activate B cells to produce IgE by inducing cytokines. The allergen-specific IgE complexes then bind to FceRI on mast cells, and the stimulated mast cells release pro-inflammatory molecules, which cause acute and chronic inflammation. Bacterial infections, mostly the result of colonization by Staphylococcus aureus or Malassezia furfur, are typically followed by dysfunction of the skin barrier, and this is usually caused by scratching.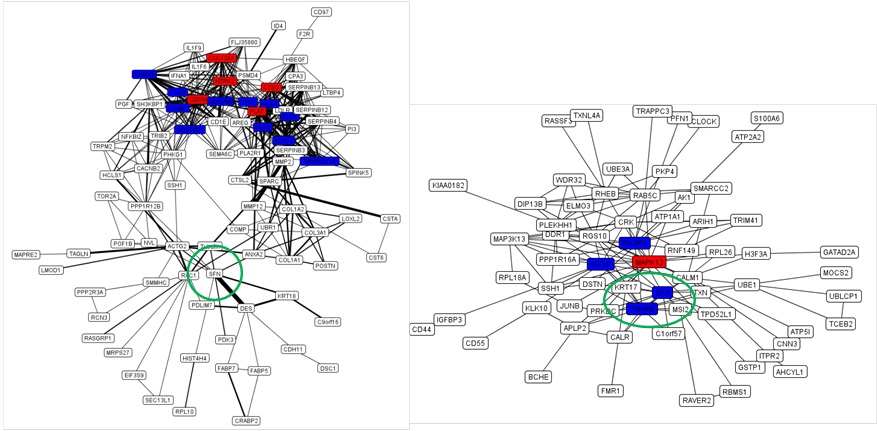
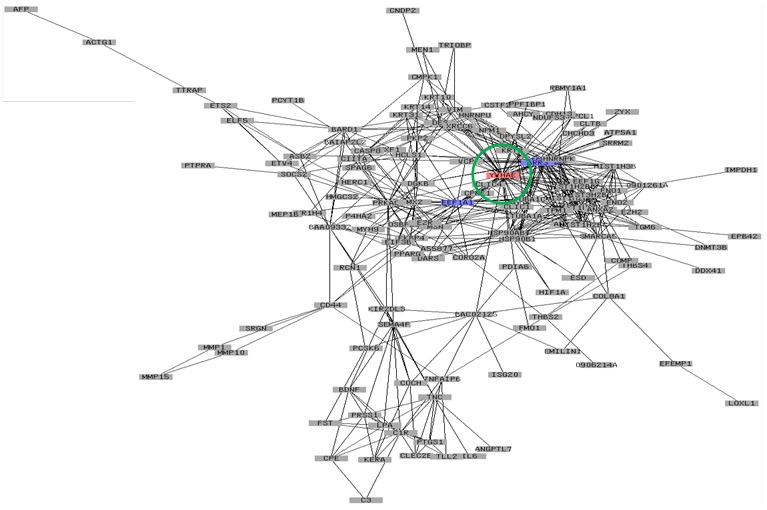
Figure 1B: The overlapping map of PEIMAP and PSIMAP from AD patient-derived fibroblasts. Labels with red and blue colors indicate the hub proteins. Red proteins are proteins that interact with more than 25% of the total proteins in the network. The blue ones are proteins that interact with between 20% and 25% of the total proteins in the network. The thickness of the edge between two proteins represents the number of reference databases. The thickness of edges is only used in PEIMAP. The data were from references [18-20], respectively.
These findings indicate that the predicted interactome can be used to analyze molecular mechanisms, not only of known genes but also of novel genes that are associated with AD pathogenesis.
SUMMARY OF AD ASSOCIATED FACTORS AND PATHOGENESIS
On the basis of previous findings, we demonstrated the pathologic process of AD and summarized its associated factors as illustrated in Scheme 1 (Figure 2). It is known that the prevalence of AD is due to complex factors such as innate genetic problems and environmental triggers. Clinically, it is known that AD simultaneously triggers asthma and allergic rhinitis for which the levels of serum IgE and peripheral eosinophils are elevated; therefore, AD is defined as a systemic disorder [24].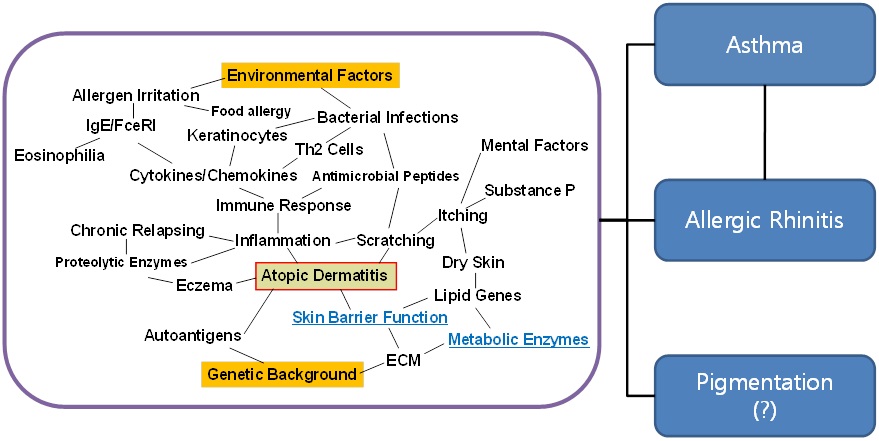
Regarding the genetic factors in AD, several candidate genes located on different chromosomes have been introduced via linkage analyses and genetic polymorphisms. SNP genetics have revealed that the associations of SNPs and haplotypes in cytokines such as IL-5, IL5RA, IL9, IL9R, IL12, IL12R, and IL-18 were strongly associated with the allergic type of AD but not with the non-allergic intrinsic type [25-28]. These data support that such cytokines upregulate IgE production to control eosinophils in the peripheral blood. The haplotypes in DEFB1 that are responsive to cutaneous colonization and infection by bacteria were significantly associated with susceptibility to AD, indicating that DEFB1 has a primary effect on skin inflammation and/or skin responsiveness to any kind of allergic reaction [29]. Analyses of the SNPs and haplotypes in SPINK5 and gene-gene interactions between SPINK5 and the DEFB1 showed associations with the susceptibility to the allergic type of AD [30]. The SNPs from DEFB1 and SPINK5 affect individual susceptibility to the development of ADE in an additive manner. Recently, the SNP in FLT4 was also found to be associated with AD [31]. These studies provide evidence for a significant interaction between allergens and suggest that SPINK5 and FLT4 may contribute to ADE susceptibility.
Regarding the immunologic aspects of AD, microbial infections in AD patients have been considered to be one of the important issues in the pathogenesis of AD, which is due to colonization by microorganisms and lipoteichoic acid, peptidoglycan, lipopolysaccharide, and S. aureus toxin B [32,33]. These infections are more seriously followed by dysfunction in both the skin barrier [34] and antimicrobial defensive peptides [35]. As mentioned previously, several blood factors associated cells in AD such as eosinophils, LCs, keratinocytes, and pro-inflammatory cells such as Th1, Th2, Th9, Th17, and Th22 have been studied extensively. Such studies found that these cells produce cytokines such as IL4, IL5, IL6, IL9, IL10, IL12, IL13, IL17, IL22, and IL31 [36-38] and chemokines such as CCL1, CCL2, CCL3, CCL4, CCL5, CCL8, CCL11, CCL13, CCL17, CCL18, CCL20, CCL22, CCL24, CCL26, CCL27 and CX3CL1 [39-41]. These factors are thought to somehow be associated with AD and play important roles in the pathway of disease occurrence. A previous study showed that Th2 cells were involved in inducing the acute stage of AD while Th1 cells could be prominent in the chronic stage of AD to sustain inflammation.
Additional external inducers such as food allergens (cow's milk, hen's eggs, wheat, and soy) and inhalant allergens (house dust mite, pollen, and animal dander) also act as provocative factors in AD, especially for children with AD [42-44]. Infants are more allergic to food, and a variety of specific allergenic responses to factors such as inhalant allergens are connected with clinical severity; infant AD has many distinctive features in its laboratory variables as compared with AD in other age groups.
Dry skin in AD usually induces itching and results in exacerbating scratching that increases the susceptibility to inflammation. For this reason, various moisturizer applications have been a basic treatment for AD patients, and some emollients are regarded as able to prevent skin inflammation in AD [45]. The cause of dry skin in AD is still unclear but it is assumed that skin barrier related genes such as Filaggrin (FLG), ceramides, sphingomyelin, and glucosylceramide associated genes are involved in dry skin when they malfunction [46-48]. Thus, stratum corneum hydration level and transepidermal water loss are laboratorial monitoring parameters for clinicians [49,50].
Mental factors are also known to be associated with itching in AD patients [51]. Psychological stress such as anxiety is thought to be associated with the induction of pruritus through mediating neuropeptides such as neuropeptide Y and substance P in AD [52,53].
Previous attempts at profiling the proteomic expression patterns of AD patient-derived cells suggested new information regarding their responses toward outward stimulation of the dermis and epidermis, including several metabolic enzymes. Metabolic enzymes are not well elucidated in AD pathogenesis; however, HTS-based research implied that their dysregulation is associated with ECM factors involved in AD for skin barrier function and lipid formation, which are correlated with the induction of dry skin.
In addition to the well-known factors for inducing AD, we have obtained more insight into AD-associated factors by OMICS studies regarding hub genes that might play central roles. Interestingly, YWHAE was previously found in melanogenesis as a hub gene [54], which implied that YWHAE is importantly related to skin pigmentation. Specifically, when the tyrosinase gene was overexpressed in HEK293 cells, it induced high pigmentation in cells. OMICS integration studies showed that YWHAE was importantly detected as a hub gene (Figure 3). We believed that skin pigmentation related with AD was not a triggering factor but might be associated with resolution or somehow involved with several important genes downstream of AD occurrence. In addition, our recent unpublished data on siRNA integrating ALDH1 downregulation in keratinocytes showed that the silencing of ALDH1 gene expression is associated with pigmentation metabolism as revealed by the gene ontology enrichment analysis. Cumulatively, this evidence all supports the association of skin pigmentation with AD disease.
FURTHER PINPOINT SPECULATION FOR AD STUDY
In further studies, there are several questions to be raised, including how to apply AD-associated factors newly identified in laboratory research into clinical applications? What are the important genes to be targeted and how can clinical output be quickly obtained using the selected target genes? Are the biomarkers or hub genes gradually revealed in AD truly important for therapeutic approaches? Before we can answer these questions, we suggest one piece of the puzzle to move AD studies forward: our significant findings concerning the ALDH1 enzyme.
In previous studies, ALDH1 (EC 1.2.1.3) was detected as being significantly downregulated in AD, and this metabolic enzyme was thought to somehow deeply associate with AD disease [55,56]. Among the profiled genes/proteins, ALDH1 could be one of the most promising target enzymes for obtaining insight into AD. To elucidate the mechanisms, our group recently conducted a siRNA OMICS integration study to identify the role of ALDH1 keratinocytes. The marked decrease in ALDH1 expression levels in both ADE and ADI patients seen in that study may provide new factors for understanding the pathogenesis of AD, which can help clarify the identities of the metabolic enzymes involved in the development of AD far beyond simple immunologic aspects. ALDH1 is the second enzyme involved in the metabolism of alcohol. The classical function of this enzyme is to catalyze the conversion of acetaldehyde into acetic acid in the alcohol metabolism pathway, along with ADH, ALDH2, and catalase. However, the isoenzyme ALDH2 actually plays a more important role than ALDH1 in acetaldehyde metabolism, predominantly in the mitochondrial matrix, while ALDH1 is cytosolic. Therefore, ALDH1 is believed to have more varied roles, which have been revealed recently: ALDH1 was proposed as a marker for cancer stem cells [57,58] and including lung cancer [59], breast cancer [60], colon cancer [61], and pancreatic cancer [62]. ALDH1 also oxidizes retinol to retinoic acid that was involved in the developing retina [63] and, also plays an important role in adipogenesis [64]. Besides, it has reported that ALDH1 appeared to be decreased in the substantia nigra in Parkinson's disease patients and also in the ventral tegmental area in schizophrenic patients [65,66]. It was suggested that the low expression levels of ALDH1 correlated with the dysfunction of the dopamine neurons. These studies appear to indicate that ALDH1 may possess additional physiological functions that are related to disease.
Obviously, if the role of ALDH1 was restricted to alcohol degradation, ALDH1 may seem less important than ALDH2; however, ALDH1 appears to function in additional pathways other than alcohol metabolism such as allergic diseases and various types of cancers. A more complete view of the many functions of ALDH1 is gradually being revealed; these functions appear to extend beyond the classical functions of ALDH1. In this regard, it is extremely interesting to consider the role of ALDH1 in skin cells, as well as the role of ALDH1 in the pathogenesis of AD. Therefore, the exact role of ALDH1 in the pathogenesis of AD and the downregulation phenomenon require further study.
CONCLUDING REMARKS
To cope with the rapidly increasing number of AD patients, making specific diagnoses and administering case sensitive treatments are important. Regardless of the various approaches, quick and specific treatment methods for AD patients are currently lacking. Therefore, HTS-based OMICS integration approaches could help researchers gain additional insights and could reveal new pieces of the AD pathogenesis puzzle. In this review we suggested new AD-associated factors based on previous reports (ALDH1, YWHAE, SFN, C1R, TNC, BARD1, SEMA4F, and LPA) as hub genes. The functions of most of these hub genes have not been well reported in AD or in the pathogenesis of other diseases. Although we did not suggest a direct role or utility of any hub gene that appears to be important in AD, we did provide hub genes to be targeted in further trials of AD diagnostic markers and/or treatment.
ACKNOWLEDGEMENTS
Dr. Jun-Mo Yang was supported by a grant from the Korea Health Technology RandD Project, Ministry of Health and Welfare, Republic of Korea (No. HI12C1299) and by a grant from the Samsung Biomedical Research Institute (GL1-B2-181-1). Dr. Yong-Doo Park was supported by a grant from Zhejiang Provincial Natural Science Foundation of China, "Towards studying the function of C3dg protein and elucidating its role in the pathogenesis of atopic dermatitis" (Grant No. LY14H110001).
REFERENCES
- Braconi D, Bernardini G, Santucci A (2010) Post-genomics and skin inflammation. Mediators Inflamm 2010.
- Blumenberg M (2012) SKINOMICS: Transcriptional Profiling in Dermatology and Skin Biology. Curr Genomics 13: 363-368.
- Leung DY (2013) New insights into atopic dermatitis: role of skin barrier and immune dysregulation. Allergol Int 62: 151-161.
- Hong X, Tsai HJ, Wang X (2009) Genetics of food allergy. Curr Opin Pediatr 21: 770-776.
- Broccardo CJ, Mahaffey S, Schwarz J, Wruck L, David G, et al. (2011) Comparative proteomic profiling of patients with atopic dermatitis based on history of eczema herpeticum infection and Staphylococcus aureus colonization. J Allergy Clin Immunol 127: 186-193, 193.
- Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA (2004) New insights into atopic dermatitis. J Clin Invest 113: 651-657.
- Elias PM, Hatano Y, Williams ML (2008) Basis for the barrier abnormality in atopic dermatitis: outside-inside-outside pathogenic mechanisms. J Allergy Clin Immunol 121: 1337-1343.
- Correale CE, Walker C, Murphy L, Craig TJ (1999) Atopic dermatitis: A review of diagnosis and treatment. Am Fam Physician 60: 1191-1198, 1209-1210.
- Schmitt J, Langan S, Deckert S, Svensson A, von Kobyletzki L, et al. (2013) Assessment of clinical signs of atopic dermatitis: a systematic review and recommendation. J Allergy Clin Immunol 132: 1337-1347.
- Tokura Y (2010) Extrinsic and intrinsic types of atopic dermatitis. J Dermatol Sci 58: 1-7.
- Wüthrich B, Schmid-Grendelmeier P (2003) The atopic eczema/dermatitis syndrome. Epidemiology, natural course, and immunology of the IgE-associated ("extrinsic") and the nonallergic ("intrinsic") AEDS. J Investig Allergol Clin Immunol 13: 1-5.
- Park JH, Choi YL, Namkung JH, Kim WS, Lee JH, et al. (2006) Characteristics of extrinsic vs. intrinsic atopic dermatitis in infancy: Correlations with laboratory variables. Br J Dermatol 155: 778-783.
- Williams HC (2005) Clinical practice. Atopic dermatitis. N Engl J Med 352: 2314-2324.
- Carr WW (2013) Topical calcineurin inhibitors for atopic dermatitis: review and treatment recommendations. Paediatr Drugs 15: 303-310.
- Krutmann J, Morita A (1999) Mechanisms of ultraviolet (UV) B and UVA phototherapy. J Investig Dermatol Symp Proc 4: 70-72.
- Park YD, Park D, Bhak J, Yang JM (2008) Proteomic approaches to the analysis of atopic dermatitis and new insights from interactomics. Proteomics Clin Appl 2: 290-300.
- Lü ZR, Park D, Lee KA, Ryu JW, Bhak J, et al. (2009) Profiling the dysregulated genes of keratinocytes in atopic dermatitis patients: cDNA microarray and interactomic analyses. J Dermatol Sci 54: 126-129.
- Lü ZR, Park TH, Lee ES, Kim KJ, Park D, et al. (2009) Dysregulated genes of extrinsic type of atopic dermatitis: 34K microarray and interactomic analyses. J Dermatol Sci 53: 146-150.
- Lü ZR, Kim WS, Cho IH, Park D, Bhak J, et al. (2009) DNA microarray analyses and interactomic predictions for atopic dermatitis. J Dermatol Sci 55: 123-125.
- Yin SJ, Si YX, Zhang M, Qian GY, Zhang F, et al. (2011) Combination of free-flow electrophoresis and interactomics to analyze the dysregulated proteins of fibroblasts from atopic dermatitis patients. J Dermatol Sci 61: 148-150.
- Ghahary A, Karimi-Busheri F, Marcoux Y, Li Y, Tredget EE, et al. (2004) Keratinocyte-releasable stratifin functions as a potent collagenase-stimulating factor in fibroblasts. J Invest Dermatol 122: 1188-1197.
- Ghaffari A, Li Y, Karami A, Ghaffari M, Tredget EE, et al. (2006) Fibroblast extracellular matrix gene expression in response to keratinocyte-releasable stratifin. J Cell Biochem 98: 383-393.
- Medina A, Ghaffari A, Kilani RT, Ghahary A (2007) The role of stratifin in fibroblast-keratinocyte interaction. Mol Cell Biochem 305: 255-264.
- Darlenski R, Kazandjieva J, Hristakieva E, Fluhr JW (2014) Atopic dermatitis as a systemic disease. Clin Dermatol 32: 409-413.
- Kim E, Lee JE, Namkung JH, Park JH, Kim S, et al. (2007) Association of the single-nucleotide polymorphism and haplotype of the interleukin 18 gene with atopic dermatitis in Koreans. Clin Exp Allergy 37: 865-871.
- Namkung JH, Lee JE, Kim E, Cho HJ, Kim S, et al. (2007) IL-5 and IL-5 receptor alpha polymorphisms are associated with atopic dermatitis in Koreans. Allergy 62: 934-942.
- Namkung JH, Lee JE, Kim E, Kim S, Kim S, et al. (2010) Association of single nucleotide polymorphisms in the IL-12 (IL-12A and B) and IL-12 receptor (IL-12Rbeta1 and beta2) genes and gene-gene interactions with atopic dermatitis in Koreans. J Dermatol Sci 57: 199-206.
- Namkung JH, Lee JE, Kim E, Park GT, Yang HS, et al. (2011) An association between IL-9 and IL-9 receptor gene polymorphisms and atopic dermatitis in a Korean population. J Dermatol Sci 62: 16-21.
- Kim E, Lee JE, Namkung JH, Kim PS, Kim S, et al. (2009) Single nucleotide polymorphisms and the haplotype in the DEFB1 gene are associated with atopic dermatitis in a Korean population. J Dermatol Sci 54: 25-30.
- Namkung JH, Lee JE, Kim E, Byun JY, Kim S, et al. (2010) Hint for association of single nucleotide polymorphisms and haplotype in SPINK5 gene with atopic dermatitis in Koreans. Exp Dermatol 19: 1048-1053.
- Namkung JH, Lee JE, Kim E, Huh IS, Park T, et al. (2013) Single nucleotide polymorphism in the FLT4 gene is associated with atopic dermatitis in Koreans. Cytokine 62: 110-114.
- Mandron M, Ariès MF, Boralevi F, Martin H, Charveron M, et al. (2008) Age-related differences in sensitivity of peripheral blood monocytes to lipopolysaccharide and Staphylococcus aureus toxin B in atopic dermatitis. J Invest Dermatol 128: 882-889.
- Matsui K, Motohashi R, Nishikawa A (2000) Cell wall components of Staphylococcus aureus induce interleukin-5 production in patients with atopic dermatitis. J Interferon Cytokine Res 20: 321-324.
- Addor FA, Aoki V (2010) Skin barrier in atopic dermatitis. An Bras Dermatol 85: 184-194.
- Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, et al. (2002) Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med 347: 1151-1160.
- Auriemma M, Vianale G, Amerio P, Reale M (2013) Cytokines and T cells in atopic dermatitis. Eur Cytokine Netw 24: 37-44.
- Yamanaka K, Mizutani H (2011) The role of cytokines/chemokines in the pathogenesis of atopic dermatitis. Curr Probl Dermatol 41: 80-92.
- Lee J, Noh G, Lee S, Youn Y, Rhim J (2012) Atopic dermatitis and cytokines: recent patents in immunoregulatory and therapeutic implications of cytokines in atopic dermatitis--part I: cytokines in atopic dermatitis. Recent Pat Inflamm Allergy Drug Discov 6: 222-247.
- Homey B, Meller S, Savinko T, Alenius H, Lauerma A (2007) Modulation of chemokines by staphylococcal superantigen in atopic dermatitis. Chem Immunol Allergy 93: 181-194.
- Julia V (2012) CX3CL1 in allergic diseases: not just a chemotactic molecule. Allergy 67: 1106-1110.
- Bao L, Zhang H, Chan LS (2013) The involvement of the JAK-STAT signaling pathway in chronic inflammatory skin disease atopic dermatitis. JAKSTAT 2: e24137.
- Werfel T, Breuer K (2004) Role of food allergy in atopic dermatitis. Curr Opin Allergy Clin Immunol 4: 379-385.
- Campbell DE (2012) Role of food allergy in childhood atopic dermatitis. J Paediatr Child Health 48: 1058-1064.
- Bergmann MM, Caubet JC, Boguniewicz M, Eigenmann PA (2013) Evaluation of food allergy in patients with atopic dermatitis. J Allergy Clin Immunol Pract 1: 22-28.
- Varothai S, Nitayavardhana S, Kulthanan K (2013) Moisturizers for patients with atopic dermatitis. Asian Pac J Allergy Immunol 31: 91-98.
- McGrath JA (2012) Profilaggrin, dry skin, and atopic dermatitis risk: size matters. J Invest Dermatol 132: 10-11.
- Murata Y, Ogata J, Higaki Y, Kawashima M, Yada Y, et al. (1996) Abnormal expression of sphingomyelin acylase in atopic dermatitis: an etiologic factor for ceramide deficiency? J Invest Dermatol 106: 1242-1249.
- Imokawa G (2009) A possible mechanism underlying the ceramide deficiency in atopic dermatitis: expression of a deacylase enzyme that cleaves the N-acyl linkage of sphingomyelin and glucosylceramide. J Dermatol Sci 55: 1-9.
- Sugarman JL, Fluhr JW, Fowler AJ, Bruckner T, Diepgen TL, et al. (2003) The objective severity assessment of atopic dermatitis score: an objective measure using permeability barrier function and stratum corneum hydration with computer-assisted estimates for extent of disease. Arch Dermatol 139: 1417-1422.
- Lynde C (2008) Moisturizers for the treatment of inflammatory skin conditions. J Drugs Dermatol 7: 1038-1043.
- Tran BW, Papoiu AD, Russoniello CV, Wang H, Patel TS, et al. (2010) Effect of itch, scratching and mental stress on autonomic nervous system function in atopic dermatitis. Acta Derm Venereol 90: 354-361.
- Oh SH, Bae BG, Park CO, Noh JY, Park IH, et al. (2010) Association of stress with symptoms of atopic dermatitis. Acta Derm Venereol 90: 582-588.
- Inagaki N, Shiraishi N, Igeta K, Nagao M, Kim JF, et al. (2010) Depletion of substance P, a mechanism for inhibition of mouse scratching behavior by tacrolimus. Eur J Pharmacol 626: 283-289.
- Lu ZR, Seo E, Yan L, Yin SJ, Si YX, et al. (2010) High-throughput integrated analyses for the tyrosinase-induced melanogenesis: microarray, proteomics and interactomics studies. J Biomol Struct Dyn 28: 259-276.
- Park YD, Lyou YJ, Yang JM (2007) Detection of down-regulated acetaldehyde dehydrogenase 1 in atopic dermatitis patients by two-dimensional electrophoresis. Exp Dermatol 16: 130-134.
- Park YD, Lyou YJ, Lee KJ, Lee DY, Yang JM (2006) Towards profiling the gene expression of fibroblasts from atopic dermatitis patients: human 8K complementary DNA microarray. Clin Exp Allergy 36: 649-657.
- Ma I, Allan AL (2011) The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev 7: 292-306.
- Douville J, Beaulieu R, Balicki D (2009) ALDH1 as a functional marker of cancer stem and progenitor cells. Stem Cells Dev 18: 17-25.
- O'Flaherty JD, Barr M, Fennell D, Richard D, Reynolds J, et al. (2012) The cancer stem-cell hypothesis: its emerging role in lung cancer biology and its relevance for future therapy. J Thorac Oncol 7: 1880-1890.
- Tan EY, Thike AA; Breast Surgical Team at Outram, Tan PH (2013) ALDH1 expression is enriched in breast cancers arising in young women but does not predict outcome. Br J Cancer 109: 109-113.
- Todaro M, Francipane MG, Medema JP, Stassi G (2010) Colon cancer stem cells: promise of targeted therapy. Gastroenterology 138: 2151-2162.
- Kahlert C, Bergmann F, Beck J, Welsch T, Mogler C, et al. (2011) Low expression of aldehyde dehydrogenase 1A1 (ALDH1A1) is a prognostic marker for poor survival in pancreatic cancer. BMC Cancer 11: 275.
- Duester G (2000) Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem 267: 4315-4324.
- Yasmeen R, Jeyakumar SM, Reichert B, Yang F, Ziouzenkova O (2012) The contribution of vitamin A to autocrine regulation of fat depots. Biochim Biophys Acta 1821: 190-197.
- Galter D, Buervenich S, Carmine A, Anvret M, Olson L (2003) ALDH1 mRNA: presence in human dopamine neurons and decreases in substantia nigra in Parkinson's disease and in the ventral tegmental area in schizophrenia. Neurobiol Dis 14: 637-647.
- Wey MC, Fernandez E, Martinez PA, Sullivan P, Goldstein DS, et al. (2012) Neurodegeneration and motor dysfunction in mice lacking cytosolic and mitochondrial aldehyde dehydrogenases: implications for Parkinson's disease. PLoS One 7: e31522.
Citation: Park YD, Yang JM (2014) OMICS Integration Studies for New Insights on Atopic Dermatitis. J Clin Dermatol Ther 1: 002.
Copyright: © 2014 Yong-Doo Park, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

