
Phylogenetic Analyses of Three Goat (Capra Aegagrus Hircus) Breeds in Cross River State, Nigeria
*Corresponding Author(s):
Edim EHDepartment Of Animal And Environmental Biology, Cross River University Of Technology, Nigeria
Tel:+080 35835981,
Email:hannahetta@crutech.edu.ng
Abstract
Phylogenetic analyses of three local goat (Capra aegagrus hircus) breeds in Calabar was carried out. This involved the isolation and extraction of DNA from twenty one blood samples collected from the 3 goat breeds - Red Sokoto (RS), Sahel White (SW) and West African Dwarf (WAD). The extracted DNA was amplified using PCR and separated in gel electrophoresis and subsequently, the gel was visualized using a UV trans-illuminator. Relatedness, genetic similarities and dissimilarities among the three goat breeds were determined using Darwin 5.0 computer software. Result showed that Red Sokoto species are genetically more distant from the WAD goat breed. The PCR amplified mtDNA loop from the 21 blood samples revealed that the ISSR primer amplified the selected genes obtained from all the blood samples. The highest number of samples were obtained from the RS breed which also showed 50% polymorphism. The evolutionary relationship among the goats sampled showed two major clusters. Cluster 1 consisted of all the RS goats and two of the SW species. Cluster 2 consisted of all the WAD goats and two of the SW goats. ISSR primers proved to be efficient in determining the genetic similarities, dissimilarities and evolutionary relatedness among the goat populations. Further investigations on the relationship between these goat breeds should be carried out towards conservation and for future breeding purposes.
Keywords
Dendrogram; Genetic distance Phylogenetic tree; Polymerase chain reaction
INTRODUCTION
Phylogenetics is the study of the evolutionary history and relationships among individuals or groups of organisms (e.g. species, or populations). These relationships are discovered through phylogenetic inference methods that evaluate observed heritable traits, such as DNA sequences or morphology under a model of evolution of these traits. The result of these analyses is a phylogeny (also known as a phylogenetic tree), a diagrammatic hypothesis about the history of the evolutionary relationships of a group of organisms [1]. Phylogenetic analyses have become central to understanding biodiversity, evolution, ecology, and genomes. Taxonomy is the identification, naming and classification of organisms. It is usually richly informed by phylogenetics, but remains a methodologically and logically distinct discipline [2]. The degree to which taxonomies depend on phylogenies (or classification depends on evolutionary development) differs depending on the school of taxonomy.
There are over 300 distinct breeds of goat [3]. Goats are one of the oldest domesticated species, and have been used for their milk, meat, hair and skin over much of the world [4]. In 2011, there were more than 924 million live goats around the globe, according to the UN Food and Agriculture Organization [5]. The West African Dwarf goat breed is from coastal west and central Africa. The West African Dwarf goat breed probably evolved in response to the conditions in the humid Forest of West Central Africa [6]. The breed is known by some other names in different parts of the world such as African Dwarf, Chevrenainede Savanes, Cameroon Dwarf, and Dwarf West Africa. There are many types of West African dwarf goats available. This goat plays a very important role in the rural economy of West Africa. Most of the poor and small scale farmers in this region raise them (Figure 1).
 Figure1: West African Dwarf, Sokoto Red, White Sahel.
Figure1: West African Dwarf, Sokoto Red, White Sahel.
The Sokoto Red goat is probably the most widespread and well-known type in Nigeria [7]. It is the usual village goat in the northern two-third of the country although it is less common with trans-humane pastoralists. The sokoto red goat was the source of “Morocco leather” known in Europe from the medieval period onwards. It acquired this name because it was transported across the Sahara by Caravans controlled by Moroccan merchants. The Sokoto Red is still known for its fine leather [8]. It has red skin coat that is of good quality for leather production. Other varieties of the breed are the Kano brown or Borno white. Both sexes carry horns with short ears that are horizontally positioned. At maturity, maradi goats weigh between 20 and 30kg. The Sahel or Desert goat is found along the northern border of Nigeria, particularly in Borno, where it is often known as ‘Balami’, although this name has not been adopted as it would lead to confusion with the better-known sheep race, Balami. [9] uses ‘Sahel’, which seems appropriate, as this race is distributed from Senegal to Sudan. In Nigeria, the Sahel goat is generally the variety preferred by pastoralists.
A close examination of the genetic distances data revealed that the smallest distance values were found between Eastern African breeds and Southern African breeds while the largest distance values were those between West African breeds and the breeds from Southern Africa. This indicates that the breeds of Southern Africa were more closely related to the Eastern African breeds than to the breeds of West Africa. The phylogeny constructed from the genetic distance data indicated that the breeds were grouped according to their geographic locations of origin and that the sub-Saharan African breeds were clearly separated from the breeds from outside Africa. A similar observation of populations clustering according to their geographic origins has been reported in humans [10] cattle [11] and chickens [12]. This implies that geographically adjacent populations are more genetically related, probably because of founder effects and interbreeding, especially around bordering areas. Bootstrap values (percentage of occurrence of a node in 1,000 bootstrap resampling of loci) ranged from 38 to 97%, with only two nodes with values below 50%. This indicates that the topology of the phylogeny constructed from DA distances was more reliable in recovering the evolutionary relationships of the populations studied. There is paucity of information on the evolutionary history of the goat breeds (West African Dwarf, Red Sokoto and Sahel White) in Calabar. The genetic differences/similarities in the goat breeds will affect future breeding programs thus there is a need to investigate and document these information. Hence this study is necessary to bring to fore, the evolutionary history and relatedness among the local goat breeds found in Calabar.
MATERIALS AND METHODS
Twenty one Blood samples were collected from three goat breeds. Twelve samples from the Red Sokoto goat (Maradi) breed which were labeled (R1-R12), four samples were taken from the White Sahel goat breed (Borno) labeled (SW1-SW4) and five samples were taken from the West African Dwarf goat breed (WAD) labeled (WD1-WD5) by an experienced veterinary laboratory technologist using sterilized 10ml syringes, to collect blood from the jugular veins of the goats. The goats were from mutton abattoirs in designated areas in Calabar, both Municipality and Calabar South. The blood collected were collected and transferred into an EDTA vacationer and mixed with anticoagulants to prevent clotting of blood and then stored at -200C until commencement of molecular analysis.
DNA Extraction: The CTAB method was used for DNA extraction. Eppendorf tubes were labeled from 1-21 according to blood samples, 20ml pipette was used to disperse 40ml of TE buffer in all tubes. 40ml pipette was used to disperse 40ml of the blood sample into the Eppend or f tubes containing TE buffer. The tubes were incubated in a water bath for 15-20minutes to destroy cells that may hinder PCR, then spun at 13000rpm using a micro centrifuge and supernatant was added to 250ul of lysis buffer, 125ul of TE buffer, 125ul of SDS and a drop of proteinase K. This was incubated for 30 minutes and centrifuged in 13000rpm for 5 minutes. 400ul of supernatant was added to 400ul of isopropanol and incubated at -30oC for 15 minutes. (Alternatively incubating at -4oC overnight). The samples were centrifuged at 13000rpm for 10 minutes. Supernatant was discarded using pipette, while pellets were left behind. Pellet were washed using 70% alcohol and centrifuged for 5 minutes removing the supernatant. The alcohol was evaporated using Centrivap concentrator. 250ul of TE buffer was added into the evaporated tubes containing the pellet. The tubes containing DNA were stored in -4oC. To check the quality of the extracted DNA samples, 1.5% agarose was weighed, dissolved in TBE (Trisma boric acid EDTA) buffer before casting the gel for electrophoresis. The gel was stained in 1.5mg/ml of ethidium bromide solution and distilled water respectively before viewing under UV trans illuminator. Some were contaminated with RNA and were later removed through the use of RNAse treatment. Following the high level of concentration of the extracted DNA samples, dilution of each DNA sample was uniformly made to 100ng/µL DNA prior to polymerase chain reaction (PCR) set up. PCR amplification was performed in volume of 25µL which consisted of 2.0µL of 50mM mgcl2 (Biolino), 2.0µL of 2.5mM dNTPs (Biolino), and 0.2µL 500µ tag DNA polymerase.(Binoline), 1.0µL DMSO (dimethlysulfoxide), 1.0µL of 10µM each primer and 16.05µof 500ml DEPC-treated water (in vitrogen corporation). The PCR cycling profile used for the reaction consisted of an initial step at 940C for 2 minutes, 40 cycles of 2minutes, and a 5-minutes final extension at 720C. The PCR reaction products were electrophoresed in a 1.5% agarose gel containing 0.5mg/ml ethidium bromide and photographed on trans illuminator UV light. DNA amplifications with each ISSR primer was repeated at least twice to ensure reproducibility. Before loading, the PCR products were first mixed with gel loading dye.The loading dye is important because it increases the density of the sample making it possible for the DNA to drop evenly into the well. For each well 2uL of sample mixed with loading buffer was used. Next to the last sample in each row, a size marker was loaded into its own well. Then 100v was applied for the migration of DNA to start and electrophoresis was allowed to continue for about 1 hour. Two ISSR primers were used for the analysis. These were (AG) 9C 5’- AGA GAG AGA GAG AGA GAG C-3’ and (GA) 9C 5’-GAG AGA GAG AGA GAG AGA C-3’. An analysis of finger print was carried out using pyElph version 1.4. The visualization of the PCR products was carried out using high performance UV trans illuminator and then photographs were taken showing samples and markers in their different lanes. Darwin 6.0 was used to generate the dendrogram and calculate the genetic distances between the goat breeds.
RESULTS AND DISCUSSION
Genetic Relatedness of the Goat Species
Figures 2 and 3 both presents the dendrogram and the phylogenetic tree respectively, showing the genetic relationships wise similarities and disimilarities among twenty one different blood samples obtained from three breeds of goats in the Calabar environs. These are the Red Sokoto (RS), the Sahel white (SW) and the West African Dwarf (WAD) goats. Twelve of the blood samples were obtained from the Red Sokoto breed (RS1-RS12), four from the Sahel white (SW1-SW4) and the remaining five from the West African Dwarf (WD1-WD5). Two main clusters emerged from the dendrogram, C1 and C2, showing the two major roots of un relatedness in the C. hircus population. The longer the distance between the clusters the higher the dissimilarities between them. The highest similarities observed are between WD5, WD3, WD 2, WD1, SW4 and SW3.
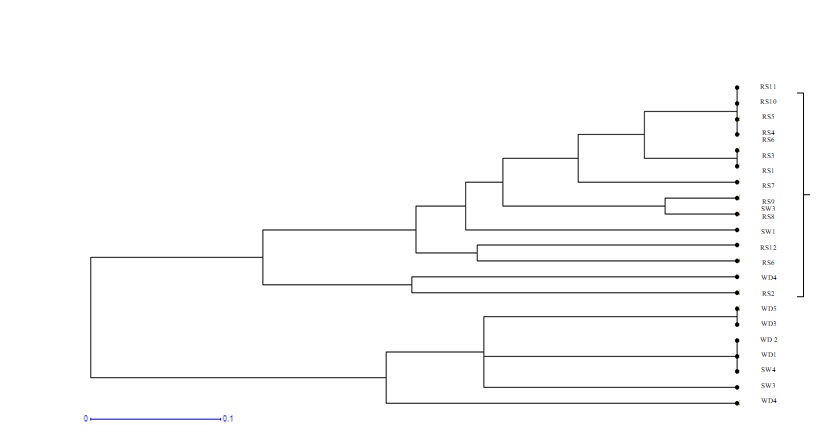 Figure 2: Dendrogram showing the Genetic relationship among the three goat breeds.
Figure 2: Dendrogram showing the Genetic relationship among the three goat breeds.

Figure 3: Phylogenetic tree showing two major clusters of the three goat breeds in Calabar environs.
Genetic Similarity and Dissimilarities among Goat Species
Table 1 presents results of the genetic distance matrix for the Red Sokoto, Sahel white and West African Dwarf breeds of goats in Calabar. The table summarizes the distances between samples based on their genetic profile. The most distantly genetically related are colored in red ink, indicating that their level of gene divergence is high. The most genetically similar species are colored in blue ink, indicating that they share ancestry and have a high genetic similarity. The distance matrix shows that RS 1 and 3, RS 4 and 5, RS 4, 10, 11, RS 5, 10 and 11, SW16, WD17 and WD18, WD 19 and WD 21 are panmictic. Interestingly, there is high genetic variability within the SW breed.
|
RS1 |
RS2 |
RS3 |
RS4 |
RS5 |
RS6 |
RS7 |
RS8 |
RS9 |
RS 10 |
RS 11 |
RS 12 |
SW 13 |
SW 14 |
SW 15 |
SW 16 |
WD 17 |
WD 18 |
WD19 |
WD 20 |
WD 21 |
|
|
RS1 |
0 |
||||||||||||||||||||
|
RS2 |
0.73 |
0 |
|||||||||||||||||||
|
RS3 |
0 |
0.73 |
0 |
||||||||||||||||||
|
RS4 |
0.14 |
0.73 |
0.14 |
0 |
|||||||||||||||||
|
RS5 |
0.14 |
0.73 |
0.14 |
0 |
0 |
||||||||||||||||
|
RS6 |
0.49 |
0.73 |
0.49 |
0.5 |
0.49 |
0 |
|||||||||||||||
|
RS7 |
0.24 |
0.73 |
0.24 |
0.2 |
0.24 |
0.49 |
0 |
||||||||||||||
|
RS8 |
0.36 |
0.73 |
0.36 |
0.4 |
0.36 |
0.49 |
0.36 |
0 |
|||||||||||||
|
RS9 |
0.36 |
0.73 |
0.36 |
0.4 |
0.36 |
0.49 |
0.36 |
0.11 |
0 |
||||||||||||
|
RS10 |
0.14 |
0.73 |
0.14 |
0 |
0 |
0.49 |
0.24 |
0.36 |
0.4 |
0 |
|||||||||||
|
RS11 |
0.14 |
0.73 |
0.14 |
0 |
0 |
0.49 |
0.24 |
0.36 |
0.4 |
0 |
0 |
||||||||||
|
RS12 |
0.49 |
0.73 |
0.49 |
0.5 |
0.49 |
0.4 |
0.49 |
0.49 |
0.5 |
0.49 |
0.49 |
0 |
|||||||||
|
SW13 |
0.42 |
0.73 |
0.42 |
0.4 |
0.42 |
0.49 |
0.42 |
0.43 |
0.4 |
0.42 |
0.42 |
0.49 |
0 |
||||||||
|
SW14 |
0.73 |
0.5 |
0.73 |
0.7 |
0.73 |
0.73 |
0.73 |
0.73 |
0.7 |
0.73 |
0.73 |
0.73 |
0.72 |
0 |
|||||||
|
SW15 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
0.99 |
0 |
||||||
|
SW16 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
0.99 |
0.39 |
0 |
|||||
|
WD17 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
0.99 |
0.39 |
0 |
0 |
||||
|
WD18 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
0.99 |
0.39 |
0 |
0 |
0 |
|||
|
WD19 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
0.99 |
0.39 |
0.39 |
0.39 |
0.39 |
0 |
||
|
WD20 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
0.99 |
0.54 |
0.54 |
0.54 |
0.54 |
0.54 |
0 |
|
|
WD21 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
0.99 |
0.39 |
0.39 |
0.39 |
0.39 |
0 |
0.54 |
0 |
Table 1: Genetic Distance Matrix among the Three Goat Breeds.
DISCUSSION
From the Cluster analysis results obtained in this study, two major clusters (Figure 2) were observed. This is similar to what was reported by [13], from their studies on genetic diversity in Gypsophila (Caryophyllaceae) species from Turkey using RAPD and ISSR analysis. Cluster 1 had samples comprising of Red Sokoto (RS 11, 10, 5, 4, 3, 1, 7, 9, 8, 12, 6 and 2) and the Sahel White (SW 14). These samples show genetic relatedness and characteristics depicting Capra hircus species while samples in the second cluster comprised of all the West African Dwarf goat breeds (WD1-WD5) and the Sahel white (SW 3 and 4) respectively. This therefore may suggest that samples in clade 1 originated from a common ancestor and that although samples in the second clade are distances apart, they all belong to a common ancestery [14] had similar results in their study. RS 11, 10, 5 and 4 belong to the same Clade or have the same Local common ancestor, RS 3 and 1 belong to the same Clade and equally originated from a Local common ancestor. RS 8 and 9 also evolved from a Local common ancestor. SW 2 and RS 2 belong to the same Clade. RS 12 and RS 6 have the same Local common ancestor. WAD 1, SW 4, WAD 4, SW 3 belong to the same clade. WAD 5 and WAD 3 belongs to the same Local common ancestor. A research conducted by [15] showed the phylogenetic dendrogram of goat breeds, with Mahabadi and Lori goat breeds grouped together, and the Markhoz breed deviating from them, This type of clustering was completely consistent with the fact that Markhoz is a mohair-producing breed but Mahabadi and Lori are known as meat and milk producing breeds.
Amplification of the mitochondrial D-loop as evident in the electrophoregram revealed that the ISSR primer amplified the selected gene obtained from all the blood samples collected from five West African Dwarf goats (WAD) which amplified within 500-600bps related to the standard DNA ladder. Four blood samples collected from the Sahel white (SW) only showed polymorphism with three out of four samples being amplified by the ISSR primers at 400-500bps of the molecular DNA ladder or barcode standard. The highest number of samples were obtained from the Red Sokoto (RS) breeds which showed 50% polymorphism. That is 6 samples out of 12 were amplified by the ISSR primers (at 300-500bps of the DNA barcode used. This research corresponds with the work of [16] who reported that approximately 700bp barcode amplicons sequenced from 30 goat samples showed a 99% similarity with Capra hircus breed Jining Quin goat mitochondrion. He therefore concluded that migration of people, trade between the people and cross breeding are the reasons for the emergence of exotic breeds in Pakistan. The study carried out by revealed that the sequenced dna showed a maximum homology (99%) with black goats (Capra hircus) and also generated DNA barcodes successfully. The results according to suggested that DNA barcodes are a highly effective characterization system for goat breeds.
Capra hircus Spp. is a preferred protein source to other kinds of meats because it is believed to be healthier. For this reason and others, there is the need for massive conservation drive of our indigenous and exotic breeds. The goat breeds used for this study - Red Sokoto (RS), Sahel White (SW), West African Dwarf (WAD) are important livestock species in the Calabar metropolis. They serve as the major mutton supply population in Calabar. The symmetrical distance matrix (Table 1), showed that the RS species (RS1-RS12) are genetically more distant from all the other samples collected. Genetic distances are used, for example, to evaluate the degree of genetic differentiation achieved during the speciation process or at other stages of evolutionary divergence [17]. Genetic distances also are used in the construction of phenograms or cladograms [18] and have indeed provided valuable information for the reconstruction of phylogenetic history on the basis of extant species. In this study, the least genetic distance (Table 1) was observed between RS 8 and RS 9 (0.11), indicating close genetic relationships between the two populations. The highest genetic distances were observed between RS 1 – RS 14 and SW 15 – SW 21(0.99), implying high genetic differentiation during the evolutionary divergence of the C. hircus populations. Genetic variation is a basic requirement for animal breeding, whereas high genetic variation is needed for the genetic improvement of domestic animals. The analysis of genetic diversity is a method to estimate the variation present in populations. ISSR was shown to be suitable and efficient in determining the genetic similarities, dissimilarities and evolutionary relatedness among the goat populations as also reported by other researchers [19,20].
CONCLUSION
The result of this investigation showed that among the samples analyzed, 50% of the Sahel breed analyzed show relatedness to both the Red Sokoto and West African Dwarf goat breed. It therefore suggests that all the samples analyzed may have some relatedness; hence the genetic diversity among the three goat breeds analyzed could be as a result of mutation, single nucleotide polymorphism and environmental factors. This study has shown that the RS species are genetically more distant apart from all the WAD goat breed, indicating that although they are seemingly related, their level of gene divergence or genetic diversity is high due to mutation and other aberrations in the chromosome and DNA. Based on the findings of this research on the genetic relatedness and similarities among the sampled goat breeds, it is recommended that West African Dwarf (WAD) goats which are of high genetic variability be utilized for breeding purposes, so as to increase heterozygosity in the goat population. It is also suggested that other genetic markers be used for investigative finger printing of these goat species.
REFERENCES
- Cain AJ, Harrison GA (2009) Phyletic Weighting. Proceedings of the Zoological Society of London 135: 1-31.
- Edwards A, Cavalli-Sforza L, Heywood V, McNeill J (1964) Reconstruction of evolutionary trees in Phenetic and Phylogenetic Classification. Online Computer Library Center Pg no: 67-76.
- Hirst K (2018) The History of the Domestication of Goats.
- Coffey L, Hale M, Wells A (2018) Goats: Sustainable Production Overview.
- Naderi S, Rezaei H, Pompanon F, Blum M, Negrini R, et al (2008) The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals. Proceedings of National Academy of Sciences 105: 17659-17664.
- Wilson RT (1991) Small Ruminant Production and the Small Ruminant Genetic Resource in Tropical Africa.Food & Agriculture Organisation Pg no: 106-114.
- Haumesser J (1975) Some aspects of reproduction in the Red Sokoto goat. Comparison with other tropical and subtropical breeds. REMVT 28: 225-233.
- Burns M (1965) The skin histology of some Nigerian goats. Tropical Agriculture (Trinidad) 42: 243–259.
- Mason IL (1988) A world dictionary of livestock breeds, types and varieties. CAB International, Wallingford, 3rd ed, UK.
- Bowcock A, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd J, et al. (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368: 455-457.
- MacHugh D, Shriver M, Loftus R, Cunningham P, Bradley D, et al. (1997) Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos Taurus and Bos Indicus). Genetics 146:1071-1086.
- Wimmers K, Ponsuksili S, Hardge T, Valle-Zarate, A, Mathur P, et al. (2000) Genetic distinctness of African, Asian and South American local chickens. Animal Genetics 31: 159-165.
- Korkmaz M, Dogan N (2015) Biogeographic pattern of genetic diversity detected by RAPD and ISSR analysis in Gypsophila (Caryophyllaceae) species from Turkey. Genet Mol Res 14: 8829-8838.
- Murital I, Afolayan O, Bemji M, Dadi O, Landi V (2015) Genetic diversity and population structure of Nigerian indigenous goat using DNA microsatellite markers. Arch Zootec 64: 93-98.
- Leila S, Alireza A, Alireza Z, Saheb F (2016) Genetic diversity and distance of Iranian goat breeds (Markhoz, Mahabadi and Lori) compared to the Beetal breed using inter-simple sequence repeat (ISSR) markers. Anim. Breed 59: 477-483.
- Ali A, Rehman A, William K (2016) Phylogenetic Analysis of Capra hircus Commonly Found Goat Breeds of Pakistan using DNA Barcode. Journal of Bioresource Management 3: 1-5.
- Askari N, Abadi M, Baghizadeh A, (2011) ISSR markers for assessing DNA polymorphism and genetic characterization of cattle, goat and sheep populations. Iranian Journal of Biotechnology 9: 222-229.
- Weir BS (1990) Genetic data analysis: Methods for discrete population genetic data, Sinauer Associates, Incorporation Publishers Pg no: 376.
- Chuanpeng N, Xiaobing W, Yanyan L, Juan Z (2012) ISSR markers as a tool for Assessing Genetic Diversity in the Chinese Alligator (Alligator sinensis). Asian Herpetological Research 3: 310-315.
- Du J, Guo HB, Li Q, Forsythe A, Chen XH, et al. (2018) Genetic diversity of Lepistanuda (Agaricales, Basidiomycota) in North east China as indicated by SRAP and ISSR Markers. Public Library of Science ONE 13: e0202761.
Citation: Edim EH, Victor IE, Kingsley NU (2020) Phylogenetic Analyses of Three Goat (Capra Aegagrus Hircus) Breeds in Cross River State, Nigeria. J Transl Sci Res 3: 010.
Copyright: © 2020 Edim EH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

