
Plant Defense Mechanisms against Bacterial Pathogens
*Corresponding Author(s):
Aqsa SaeedDepartment Of Plant Breeding And Genetics, PMAS Arid Agriculture University Rawalpindi, Pakistan
Email:aqsasaeed1996@gmail.com
Abstract
Plants were successful in developing complicated defense mechanisms against pathogen infections during the process of evolution. Though the underlying molecular mechanisms behind this immunity are still not very well understood. Globally, the diseases which are caused by pathogens in significant crops are negatively and greatly impacting the production of agriculture and the environment. The interactions between plants and bacteria are highly intricate because of numerous plant-signalling events occur and bacterial factors are present, which eventually describe the resistance or susceptibility of the plant liable to pathogen. Therefore, it is important to better recognise the mechanisms through which plants use to resist the infection of bacteria or to establish an effective defense response, as a better and thorough apprehension of the molecular mechanisms of such interactions will ultimately confer a transgenic approach, to minimize the impact of most of the devastating plant diseases. Additionally, the insights from the recent omics approaches will ultimately influence our enlightment of the cellular and molecular procedures governing host responses like ETI, ETS and PTI.
Keywords
Bacterial pathogens; ETI; Plant Defense; PAMP-Triggered Immunity (PTI)
ABBREVIATIONS
RNA, Ribonucleic Acid; RBPs, RNA-Binding Proteins; PRRs, Protein Recognition Receptors; PAMPs, Pathogen-Associated Molecular Pattern Molecules; ETI, Effector-Triggered Immunity; PTI, PAMP-Triggered Immunity; MAMPs, Microbe-Associated Molecular Pattern Molecules; ETS, Effector-Triggered Susceptibility; LRR, Leucine-Rich Repeat; NB, Nucleotide-Binding; PR, Pathogenesis-Related; SA, Salicylic Acid; HR, Hypersensitive Response; PCD, Programmed Cell Death; HSTs, Host Selective Toxins; JA, Jasmonic Acid; DNA, Deoxyribonucleic Acid; R-genes, Resistance genes; PA, Plant Associated; EFTu, Elongation Factor Thermos Unstable; Avr, Avirulence; SAR, Systemic Acquired Resistance
INTRODUCTION
Plants were successful in developing complicated defense mechanisms against pathogen infections during the process of evolution. Jeffery Dangl and Jonathan Jones gave the term ‘Plant immune system’ in 2006, which relates with the elaborative plant defense system of plants against phytopathogenic microbes [1]. Its efficiency and advancement in order to suppress the pathogenic infections vary amid plant species. It has been noted that the medicinal plant immune is perhaps better established, and it is shown by their efficient capability of protection against numerous pathogens. Though the underlying molecular mechanisms behind this immunity are still not very well understood. Particularly, very less is identified about the RBPs or RNA-Binding Proteins which exist in medicinal plants primarily because of the inadequate amount of identified RBPs, yet, CmGRP1, are cognized RBP, indicates probable significance of RBPs in the immunity of the plant is present in sap of a medicinal plant called Chelidonium majus. [2,3].
Mostly the defence in plants is initiated when a specific molecule of pathogen or one of its structural characteristics is being familiarized by the transmembrane PRRs on the surface of the cell of a plant. This pattern of recognition depends upon the conserved molecular characteristics of fungal or bacterial source, which is pathogen-associated o Microbial Associated or Molecular Patterns (MAMPs or PAMPs). It will lead to initiation of defense genes expression and PAMP-Triggered Immunity (PTI), which ultimately avert pathogenesis. Although pathogens might discharge effect or molecules and exceed PTI leading towards Effector-Triggered Susceptibility (ETS). Additionally, plants also have resistance proteins which normally contain Leucine-Rich Repeat (LRR) and Nucleotide-Binding (NB) domains triggering signalling cascade after identifying exact effectors. Hence, ultimately leading towards triggering the expression of downstream genes in order to create a fast and robust defense response which prevents the spread of pathogens. This triggered defense response and the acknowledgement of effector molecules by R proteins are collectively termed Effector-Triggered Immunity (ETI). Then received signals from the recognition of effector are then communicated to nucleus for complementing gene expression related with defense. They may also code for transcription factors for the initiation of transcription of downstream enzymes which is mandatory to produce metabolites which are related with defense like Pathogenesis-Related (PR) or Salicylic Acid (SA) proteins. Such transduction of signals leads towards Hypersensitive Response (HR), and is categorised for the synthesis of PR proteins, accumulation of Salicylic acid and reactive oxygen species. This HR eventually leads to programmed cell death or PCD for inhibiting invasion by pathogen [4].
Finally, the existing plant immunity model describes the responses of ETI happen more swiftly; they are lengthier, and also sturdier than the responses of PTI, signifying the fact that PTI is a frailer variant of ETI [1]. Therefore, typically the association of ETI with an SAR and HR in the absence of PTI. However, there are certain PAMPs like HrpZ harp in which have the capability for inducing hypersensitive response in plants [5], representing that hypersensitive response induction can happen in other responses dissimilar from ETI. Moreover, it is also known that PAMPs may also trigger systemic acquired resistance in the plants [6]. While contrary to it, some unusual examples of feeble responses of ETI were also been observed along with an effector protein [7]. So, by considering all the above-mentioned deliberations, it was recommended that the difference among effectors and PAMPs, among resistance proteins and PAMP receptors, hence also among ETI and PTI cannot be sustained firmly [8]. In its place a hypothesis was proposed that there exists a continuum among ETI and PTI, which depicts that resistance in plant, is determined through immune receptors that identify suitable legends for the activation of defense, largeness of which is probably determined through essentiality of operative immunity [8].
Globally, pathogen caused diseases in vital crops have a significant negative impact on environment and on production of agriculture. Therefore, a better understanding of mechanisms utilized by plants for resisting the infection by bacteria or an effective defense response is important, as a deeper and extra thorough recognition of the molecular mechanism of such interactions will ultimately allow transgenic methods to play their role significantly for the reduction of effects of most of the distressing plant diseases.
STRATEGIES UTILIZED BY BACTERIAL PATHOGENS TO CONQUER PLANT DEFENSE
Plants have successfully evolved numerous highly and generally specified defense mechanisms whose purpose is the prevention of diseases which are caused by wide number of microbial pathogens they come across. These defense mechanisms comprise induced and preformed antimicrobial compounds to deter attack of pathogen, cell wall strengthening to avert the entrance of pathogen, and (PCD) or programmed cell death to restrict pathogen spread and establishment. When the development of disease occurs, virulent pathogen generally infects preferably a specific plant cultivar or species, which suggests the presence of highly evolved specified tactics for the promotion of disease. It’s been hypothesized very earlier that effective pathogens rely, at least in part, on their capability to actively suppress or avoid the defense response of plant [9,10]. Therefore, effective disease development may depend on the factors of the pathogen that serve for inducing susceptibility into an otherwise tolerant or resistant host. Various pathogen city factors have been acknowledged, including (HSTs) or host selective toxins and small molecule suppressors from phytopathogenic fungi, type III effectors and toxins from phytopathogenic bacteria, and suppressors of post-transcriptional gene silencing from plant viruses [11-15].
However, the mechanisms utilized by bacteria for the suppression of plant immunity are generally not characterized. Although some discoveries have revealed the use of various strategies by bacteria to undermine the defense mechanism of plants and target the central components of the immunity of plant, such as cell wall defenses, PCD built on Hypersensitive Response (HR), and Jasmonic Acid (JA) signalling. Therefore, the identification of plant defense mechanisms that may be attacked by pathogens has directed to an improved understanding of the essential processes for both plant immunity and bacterial pathogenesis. These findings therefore present an excessive potential for improving plant resistance to disease.
PROTEOMICS APPROACH FOR UNRAVELLING PLANT RESPONSE TO BACTERIAL PATHOGENS
Continuously plants are prone to microbes and they respond them by the activation of their inbuilt immune response, including race-specific and basal resistance mechanisms [1]. Thus, to obtain a detailed knowledge of the defense systems of plant, the recognition of the compound and diverse signalling cascades and the numerous interacting biochemical pathways that are triggered by pathogen is therefore a necessity. Numerous studies on this complication using DNA microarrays or microarrays have previously been performed to examine the overall transcriptome changes caused by pathogens [16-18]. However, the transcriptional modifications represent only part of the answer because they do not provide information on post-translational and post-transcriptional processes, protein turnover and activation.
This important data can be obtained by the approach of proteomics, which permits the differences in abundance of protein which exist while sampling to be monitored and provides a space to study the pathways of communication between organelles and protein trafficking. Thus, in order to have a clearer view of the elements regulating plant-pathogen interactions, certain complementary methods like the proteome-based expression profiling are required. Here, the focus will be on few of the latest advances in the utilization of proteomics for the improvement of the understanding of specific host and host defense responses caused by bacterial pathogens [19-21]. The material of plant used in proteomic studies includes stems, roots and leaves and also the suspensions and callosities. Inoculation of the tissue of a plant is done with the pathogenic bacteria or by using the components of bacteria that constitute elicitors or PAMP. The effectors cause diseases in susceptible plants or they trigger an R-gene-mediated EIT when a bacterial PAMP generates a plant defense response. After inoculation, the sample can be evaluated at different times. The obtained protein samples as well as the suitable biological replicates are separated by two-Dimensional gel Electrophoresis (2-DE), which is usually trailed by staining of the proteins. The most commonly used protein stains are Sypro dyes, deep purple, Coomassie brilliant blue, silver nitrate, ProQEmmerald, immunoassay and CyDyes. The relatively less common methods are radioactive isotopes and Zn negative staining. Before choosing the detection method, some essential attributes such as dynamic range, reproducibility, linearity, sensitivity, compatibility with identification methods, and protein to protein variability should be taken into account [22,23].
Therefore, the use of proteomic approaches and their ideas will ultimately lead to an important enlightment of the cellular and molecular processes that administer replies of host for example FTA, ETI and PTI. In addition, the overall worldwide evaluation of the response of different pathways utilizing the approach of proteomics allowed us to identify new proteins whose biological role allows a thorough cellular and molecular elucidation. Lastly, a system-level understanding of these biotic stress responses will lead us to identify capable new targets for the purpose of developing improved cultivars.
BACTERIAL ADAPTATION TO PLANTS A GENOMIC POINT OF VIEW
The micro biota of plants and animals has evolved simultaneously along with their host families for millions of years [24-26]. It is because of the process of photosynthesis that plants are a potential carbon source for varied communities of bacteria. This comprises commensal and mutualists, as well as pathogens. Plant growth-promoting bacteria and Phytopathogens significantly disturb the health of plants and its growth and productivity [27-30]. Except for the majorly studied relationships like T-DNA transfer by Agrobacterium, type III secretion-mediated pathogenesis and nodulation of roots in legumes, the understanding of molecular mechanisms leading the plant-microbe interactions is quite incomplete [31-33]. So, the identification and characterization of bacterial functions and genes helping them to thrive in the environment of plant is a necessity. Such kind of knowledge enhance our ability to fight with diseases of plant and harness advantageous bacterial attributes for agriculture, which will directly impact bioenergy, carbon sequestration and food security.
Culture-independent methods based on shotgun metagenome sequencing or marker gene profiling has significantly improved our understanding of microbial ecology importance in the plant environment [34-38]. At the same time, reduced costs of sequencing have also permitted the genome sequencing of Plant-Associated bacterial isolates (PA) on a larger scale [39]. In addition, isolates also allow functional validation of in silico forecasts. In addition, the isolation genome provides an evolutionary and genomic background for individual genes and the ability to penetrate the genomes of unique organisms that may have been unexploited by metagenomics due to the limited depth of sequencing. Even though metagenome sequencing possesses the benefit of acquiring the DNA from non-cultured organisms, multiple 16S rRNA gene runs have shown reproducibly that the most prevalent plant-associated bacteria are primarily a derivative of four phyla. [34,35,38-40] (Actinobacteria, Proteobacteria, Firmicutes and Bacteroidetes) that can be grown. Therefore, bacterial culture is not a main obstacle at the time of sampling most members of the microbiome of the plant [39]. The part of plant-associated microbial communities in the health and growth of the host is also increasing. Therefore, a genomic level understanding of plant-microbe relationships could help enrich agricultural productivity with microbes. Many studies have emphasized on precise plant microbiomes, with a greater stress on microbial diversity rather than gene function [36,37,39,41-48].
Therefore, it has been concluded that many PA functions are surprisingly constant among phylogenetically varied bacterial taxons and that some attributes are even common with the eukaryotes of PA. In addition, these traits could enable the colonization of plants by microbes and could prove to be beneficial in agricultural inoculants genome engineering in order to ultimately produce more sustainable and more efficient agriculture (Figure 1).
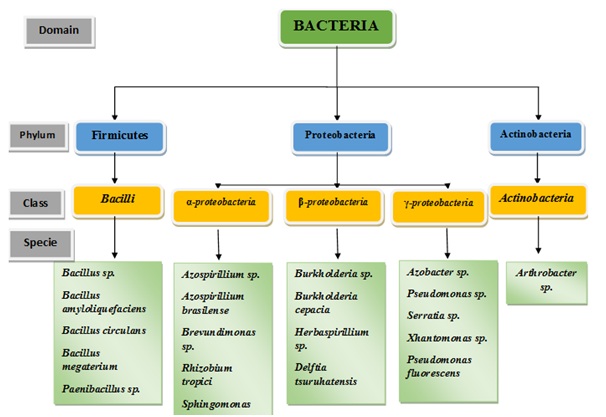 Figure 1: The most frequently grouped and studied PGPR according to their Phylogenetic classification.
Figure 1: The most frequently grouped and studied PGPR according to their Phylogenetic classification.
AN OVERLAP BETWEEN GENE-FOR-GENE AND INNATE DEFENSE RESPONSE
The outer surface of the plant has a waxy cuticle that has antimicrobial properties to avoid the intrusion of numerous invaders. The cell wall provides intruders that gain access to indoor spaces an operative second barrier. However, any intruder that overcomes these two barriers must overcome the difficult task of overpowering the immune response of plant. The immunity of a plant can be divided into two factors that work on diverse time scales. Moreover, the appearance of basal defense system is first in the pathogen interaction, whereas the Resistance gene (R) mediated defense works on the hourly time scale.
The mediation of the early basal response is done by PAMPs containing peptidoglycan, lipopolysaccharide, mannan of yeast, and bacterial flagellin [49]. It has also been found that the elongation factor of bacteria, EFTu which induces the innate response of immune system [50]. As these compounds are not only pathogens associated but also occur in non-pathogens, the term “pathogen-associated” is a misnomer [51]. Recognition of PAMPs occurs through the receptors positioned within the plasma membrane and then activates a phosphorylation cascade during binding that leads to the induction of early basal resistance, which plays its role in preventing it colonization by non-pathogenic bacteria [52]. Characteristically, the PAMP-elicited immunity is sufficient to stop the infection prior the microbes settle [53]. Therefore, an association between the reduction of growth of the pathogen and recognition of PAMP flagellin via the receptor FLS2 has been established [54].
It has been shown that flagellin, which is a major flagellar protein and also a characterized PAMP, is known from the Leu-rich repeat receptor kinase FLS2 in Arabidopsis. FLS2 is in the plasma membrane and is supposed to be involved in the early bacterial and plant interaction through its recognition and binding to flagellin. The bacterial effector proteins are a series of bacterial proteins that are shown to be involved in the deactivation of host defense systems. Some effector proteins of bacteria are Avirulence factors (Avr factors) which cooperate with host R proteins as a consequence of gene-for-gene interaction [53,55].
Pathogens are involved in secretion of DNA or effector proteins into the host cell to try to overpower the plant defense system, an operative virulence strategy of plant pathogens. Secretion system is of three typesin phytopathogenic bacteria. Type II, which is present in the genus Erwinia and is utilized for the secretion of cell wall degrading enzymes which cause soft rot, whereas type IV transfers DNA and proteins from Agrobacterium. There is also a third type, type III (T3SS), which is characteristic of Pseudomonas pathovars and excretes effector proteins in the plant cells [56]. T3SS is a multiprotein complex associated with the bacterial flagellum. Moreover, in T3SS, the bacteria in the apoplastic space via a pilus for the injection of effector proteins into the plant cells. Effector proteins are also secreted by fungal pathogens, possibly via the haustoria, but not in the cell, but in the apoplast.
Therefore, it can be concluded that gene-for-gene-mediated defense is an inherited one and limited to a specified pathogen. Plants possess dominant R genes, the product of which enables the identification of the Avr allele’s complementary alleles. Avr proteins are effector proteins that are secreted into the plant cell to promote the virulence of the pathogens and overpower the host's defenses. Localized programmed cell death and the hypersensitive response are a trademark of R-gene-mediated defense and also a target of effector proteins.
CONCLUSION
Plants and bacterial interactions are severely complicated as there are numerous factors of bacteria and signalling events of plants that ultimately determine the resistance or susceptibility of the plant which is visible to the pathogen. Many latest studies focus on improved understanding of molecular mechanisms associated with specific and basal defense responses of plants towards bacterial pathogens. However, as further discrete details of the immunity of plant are dissolving, the contributions of the gene-for-gene-mediated responses and basal responses to bacterial pathogens are gradually becoming cleared and the dissimilarity among them is revealing. In addition, the findings of recent omics approaches will ultimately influence our enlightment of the cellular and molecular processes driving the responses of host like ETI, ETS and PTI. Finally, systemic biotic stress response understanding will be beneficial for the identification of new targets in order to develop new varieties with enhanced resistance of disease.
AUTHOR CONTRIBUTIONS
AS came with idea and wrote the manuscript while T, critically reviewed and proofread the manuscript.
CONFLICT OF INTEREST
All authors declare no conflict of interest.
REFERENCES
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323-329.
- Nawrot R, Kalinowski A, Gozdzicka-Jozefiak A (2007) Proteomic analysis of Chelidonium majus milky sap using two-dimensional gel electrophoresis and tandem mass spectrometry. Phytochemistry 68: 1612-1622.
- Nawrot R, Tomaszewski ?, Czerwoniec A, Go?dzicka-Józefiak A (2013) Identification of a Coding Sequence and Structure Modeling of a Glycine-Rich RNA-Binding Protein (CmGRP1) from Chelidonium majus L. Plant Mol Biol Rep 31: 470-476.
- Woloshen V, Huang S, Li X (2011) RNA-binding Proteins in Plant Immunity. J Pathog 2011: 278697.
- Lee J, Klüsener B, Tsiamis G, Stevens C, Neyt C, et al. (2001) HrpZPsph from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc Natl Acad Sci U S A 98: 289-294.
- Mishina TE, Zeier J (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50: 500-513.
- Wirthmueller L, Zhang Y, Jones JD, Parker JE (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol 17: 2023-2029.
- Thomma BP, Nürnberger T, Joosten MH (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4-15.
- Bushnell WR, Rowell JB (1981) Suppressors of defense reactions: A model for roles in specificity. Phytopathology 71: 1012-1014.
- Heath MC (1981) A generalized concept of host-parasite specificity. Phytopathology 71: 1121-1123.
- Wolpert TJ, Dunkle LD, Ciuffetti LM (2002) Host-selective toxins and avirulence determinants: what’s in a name? Annu Rev Phytopathol 40: 251-285.
- Shiraishi T, Yamada T, Ichinose Y, Kiba A, Toyoda K (1997) The Role of Suppressors in Determining Host-Parasite Specificities in Plant Cells. Int Rev Cytol 172: 55-93.
- Collmer A, Lindeberg M, Petnicki-Ocwieja T, Schneider DJ, Alfano JR (2002) Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol 10: 462-469.
- Bender CL, Alarcon-Chaidez F, Gross DC (1999) Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63: 266-292.
- Moissiard G, Voinnet O (2004) Viral suppression of RNA silencing in plants. Mol Plant Pathol 5: 71-82.
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, et al. (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiology 135: 1113-1128.
- Truman W, de Zabala MT, Grant M (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J 46: 14-33.
- Cernadas RA, Camillo LR, Benedetti CE (2008) Transcriptional analysis of the sweet orange interaction with the citrus canker pathogens Xanthomonas axonopodis pv. citri and Xanthomonas axonopodis pv. aurantifolii. Mol Plant Pathol 9: 609-631.
- Mehta A, Brasileiro AC, Souza DS, Romano E, Campos MA, et al. (2008) Plant-pathogen interactions: what is proteomics telling us? FEBS J 275: 3731-3746.
- Mathesius U (2009) Comparative proteomic studies of root-microbe interactions. J Proteomics 72: 353-366.
- Cheng Z, Woody OZ, Glick BR, McConkey BJ (2010) Characterization of plant-bacterial interactions using proteomic approaches. Current Proteomics 7: 244-257.
- Westermeier R, Marouga R (2005) Protein detection methods in proteomics research. Biosci Rep 25: 19-32.
- Miller I, Crawford J, Gianazza E (2006) Protein stains for proteomic applications: which, when, why? Proteomics 6: 5385-5408.
- Baumann P (2005) Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59: 155-189.
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, et al. (2008) Evolution of mammals and their gut microbes. Science 320: 1647-1651.
- Sprent JI (2008) 60Ma of legume nodulation. What’s new? What’s changing? J Exp Bot 59: 1081-1084.
- Fibach-Paldi S, Burdman S, Okon Y (2012) Key physiological properties contributing to rhizosphere adaptation and plant growth promotion abilities of Azospirillum brasilense. FEMS Microbiol Lett 326: 99-108.
- Chowdhury SP, Hartmann A, Gao X, Borriss R (2015) Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review. Front Microbiol 6: 780.
- Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, et al. (2015) Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci USA 112: 5013-5020.
- Pfeilmeier S, Caly DL, Malone JG (2016) Bacterial pathogenesis of plants: future challenges from a microbial perspective: Challenges in Bacterial Molecular Plant Pathology. Mol Plant Pathol 17: 1298-1313.
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271-282.
- Hueck CJ (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62: 379-433.
- Peters NK, Frost JW, Long SR (1986) A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science. 233: 977-980.
- Gottel NR, Castro HF, Kerley M, Yang Z, Pelletier DA, et al. (2011) Distinct Microbial Communities within the Endosphere and Rhizosphere of Populus deltoides Roots across Contrasting Soil Types. Appl Environ Microbiol 77: 5934-5944.
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91-95.
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, et al. (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86-90.
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, et al. (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64: 807-638.
- Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, et al. (2014) Niche and host-associated functional signatures of the root surface microbiome. Nat Commun 5: 4950.
- Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, et al. (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528: 364-369.
- Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, et al. (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79: 293-320.
- Shade A, McManus PS, Handelsman J (2013) Unexpected diversity during community succession in the apple flower microbiome. MBio 4: 602-612.
- Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, et al. (2013) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 7: 2248-2258.
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, et al. (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110: 6548-6553.
- Bokulich NA, Thorngate JH, Richardson PM, Mills DA (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci USA 111: 139-148.
- Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, et al. (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA 112: 911-120.
- Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, et al. (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17: 392-403.
- Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, et al. (2016) Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLOS Biol 14: 1002352.
- Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, et al. (2016) Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209: 798-811.
- Nurnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198: 249-266.
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, et al.(2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496-3507.
- Ausubel FM (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6: 973-979.
- Go´mez-Go´mez L, Boller T (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci 7: 251-256.
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803-814.
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, et al. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764-767.
- Dangl JL, McDowell JM (2006) Two modes of pathogen recognition by plants. Proc Natl Acad Sci USA 103: 8575-8576.
- Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7: 601-611.
Citation: Saeed A, Tahira (2019) Plant Defense Mechanisms against Bacterial Pathogens. J Genet Genomic Sci 3: 012.
Copyright: © 2019 Aqsa Saeed, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

