
Predicting the mutagenic effects of variants rs7188856 and rs7110 of N-Acetylglucosamine-1-Phosphodiester Alpha-N-Acetylglucosaminidase (NAGPA) gene detected in randomly collected blood samples
*Corresponding Author(s):
Santosh MaruthyDepartment Of Speech-Language Sciences, All India Institute Of Speech And Hearing, Naimisham Campus, Manasagangothri, Mysrur, India
Tel:+91 9663922033,
Email:santoshm@aiishmysore.in
Abstract
N-Acetylglucosamine-1-Phosphodiester Alpha-N-Acetylglucosaminidase (NAGPA) gene codes for Uncovering Enzyme (UCE). UCE catalyses the addition of mannose-6-phosphate as a recognition tag on lysosomal enzymes directed to lysosome. Mutations in NAGPA are known to cause stuttering, a common speech disorder with unknown aetiology. Currently, variants of three genes GNPTAB [NM_024312.4] (encoding N-acetylglucosamine-1-phosphotransferase alpha/beta subunits precursor), GNPTG [NM_032520.4] (encoding N-acetylglucosamine-1-phosphotransferase gamma subunit) and NAGPA [NM_016256.3] (encoding N-acetylglucosamine-1-phosphodiester alpha-N-Acetylglucosaminidase) have been associated with non-syndromic stuttering in populations from United States, England, Pakistan, Cameroon, Brazil and India. In the present study, 80 participant’s blood samples with no stuttering history collected and subjected to Ion proton NGS analysis revealed two major variants rs7188856 at position T465I and rs7110 at 3’UTR region in NAGPA gene. As the information regarding to their biological role not available we subjected to bioinformatic analysis in order to evaluate its mutagenic effects that would help in preforming biological assays and implications on the pathway targeting hydrolytic enzymes to lysosomes which is linked to stuttering and other disorders. The dynamut2 analysis of Single Nucleotide Variant (SNV) rs7188856 revealed that the missense mutation at the position 465 with replacement of Threonine to Isoleucine in the current predicted NAGPA 3D structure will causes destabilization and the ΔΔG stability was brought down to -0.46 Kcal/mol. The RNA fold webserver output for the SNV rs7110 revealed that the positions of loops and stems in both the structures are almost same except at the variant site. There is a change in the position of loop and stem in the predicted secondary 3’UTR NAGPA structure. We also performed the scanning of 3’UTR region containing rs7110 mutation but it did not result any miRNA target site. Therefore, though final confirmation of variant-effect predictions has to come from experiments and computational methods suggest what experiments to be performed.
Keywords
NAGPA; Variants; rs7188856; rs7110; stuttering; NGS
Introduction
N-Acetylglucosamine-1-Phosphodiester Alpha-N-Acetylglucosaminidase (NAGPA) gene codes for Uncovering Enzyme (UCE). UCE catalyses the addition of mannose-6-phosphate as a recognition tag on lysosomal enzymes directed to lysosome. The human NAGPA gene is transcribed into two different forms, probably due to alternative splicing. One of them, known as a brain isoform, is lacking exon 8 (102bp) a study suggests that the cerebral cortex, expressing the highest quantity of the NAGPA brain isoform, might be the region associated with speech function [1]. Mutations in NAGPA are known to cause stuttering, a common speech disorder with unknown aetiology. Stuttering is a disorder related to brain development identified as interruptions in normal course of speech in the form of repetitions, prolongations and involuntary pauses. The secondary behaviours synced with stuttering are eye blinking, jaw jerking, and head movements. It is believed that these secondary movements happen as an effort to decrease the severity of the stuttering. However, little is known about genetic basis of stuttering to date. Unknown pattern of inheritance and multifactorial nature of stuttering have made it difficult to find responsible genetic alterations. Currently, variants of three genes GNPTAB [NM_024312.4] (encoding N-acetylglucosamine-1-phosphotransferase alpha/beta subunits precursor), GNPTG [NM_032520.4] (encoding N-acetylglucosamine-1-phosphotransferase gamma subunit) and NAGPA [NM_016256.3] (encoding N-Acetylglucosamine-1-Phosphodiester Alpha-N-acetylglucosaminidase) have been associated with non-syndromic stuttering in populations from United States, England, Pakistan, Cameroon, Brazil and India [2-4]. According to previous study 6-18% of stuttering cases found to be associated with variants of these genes [3]. In the present study, blood samples of 80 participants with no stuttering history collected and subjected to Ion proton NGS analysis revealed two major variants rs7188856 at position T465I and rs7110 in 3’UTR region in NAGPA gene. As the information regarding to the biological role not available we subjected NAGPA variants to bioinformatic analysis in order to evaluate their mutagenic effects and to predict the effect on the protein-protein interactions that would help in preforming biological assays and implications on the pathway targeting hydrolytic enzymes to lysosomes which is linked to stuttering and other disorders.
Materials and Methods
Selection of study participants
The participants (n = 80) included in the current study were non-stutters. All the study procedures were adhered to the principles of the Declaration of Helsinki and were approved by the Institutional Ethical Committee. Written informed consent was obtained from the participants.
Sample collection, DNA isolation and NGS analysis
About 5 ml of peripheral venous blood was collected from the study participants (n =80). DNA was extracted through Proteinase-K digestion followed by phenol–chloroform extraction and ethanol precipitation. After quantification approximately 100ng of genomic DNA was used to construct Exome libraries using Ion Ampliseq Exome RDY Panel (Thermo Fisher Scientific) as per the manufacturer’s protocol and these were quantified using Qubit 3.0 (Thermo Fisher Scientific). Approximately 25 Pico moles of the library was used for template generation, enrichment and chip loading following user guidelines. Sequencing was performed using Hi-Q chemistry on Ion Proton system (Thermo Fisher Scientific) at our facility.
Unaligned binary data files (Binary Alignment Map, BAM) generated by the Ion Proton system were uploaded to Ion Reporter software (Thermo Fisher Scientific) and analysed using default settings. Sequences were aligned against the reference genome (GRCh38/hg19) by using TMAP Alignment (Thermo Fisher Scientific).
Bioinformatic analysis
The 3D protein structure of NAGPA: Complete DNA and Protein sequences of NAGPA gene were downloaded from the NCBI website. The 3D structure was not available in the Protein Data Bank (www.rcsb.org). In order to get complete3D structure we searched for other websites and predicted 3D structure was found in AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk) with an accession number Q9UK23. The structure file was downloaded and used for further analysis.
Structure of 3’UTR region of NAGPA gene: The 3’UTR region of NAGPA gene which is about 634 base long was downloaded from NCBI’s ACE VIEW website (www.ncbi.nlm.nih.gov/ieb/research/acembly/index.html). Prior to structure prediction the DNA sequence was further transcribed into RNA sequence through Biomodel (http://biomodel.uah.es/en/lab/cybertory/analysis/trans.htm). The 3’UTR structure of both wild type and mutant type were designed through RNA fold webserver (rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi). The best fit with least energy was selected for further analysis.
Predicting mutation/damaging effect of SNP rs7188856 and rs7110: The NAGPA protein structure and site of the mutation (T465I) both were submitted to DynaMut2 (http://biosig.unimelb.edu.au/dynamut2) to compare with wild type and mutant structure in order to predict the effect of mutation at level of protein 3D structure.Similarly, another mutation rs7110 in 3’UTR region was further subjected to bioinformatic analysis. The wild and mutant UTR structures were compared to each other in order to see the difference in the structure after change in the nucleotide base was observed. Most often a UTR mutation creates a microRNA target site and responsible for down regulation of the respective gene [5]. We scanned 3’UTR region containing rs7110 mutation for any miRNA site through targetscan [6] and miRanda [7].
Results
NGS data analysis
The Ion Reporter software (Thermo Fisher Scientific) displayed two variants of significantly important are rs7188856 and rs7110 both belong to NAGPA gene in the current study. The rs7188856 variant arises due to change in the amino acid from Threonine to Isoleucine at position 465 (T465I). The other mutation was found on the 3’UTR region and the nucleotide change observed was from “G” to “A”. The rs7188856 variant appeared in the current study with frequency of 24 % and rs7110 with a frequency of 100 %.
Bioinformatic analysis
The 3D protein structure of NAGPA: The protein structure was downloaded from AlphaFold Protein Structure Database (Figure 1) to further carryout the mutagenic pathogenic effect analysis.
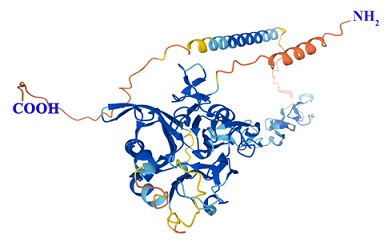 Figure 1: The Alfa fold predicted 3D structure of Human NAGPA Protein with an accession number Q9UK23 (Source:https://alphafold.ebi.ac.uk).
Figure 1: The Alfa fold predicted 3D structure of Human NAGPA Protein with an accession number Q9UK23 (Source:https://alphafold.ebi.ac.uk).
Mutagenic effect of rs7188856: DynaMut2, a server based utilityworks on Normal Mode Analysis (NMA) methods to capture protein motion and representing wild type environment investigates the effects of single and multiple point mutations on protein stability and dynamics. DynaMut2 predicts the effects of missense mutations on protein stability, achieving Pearson's correlation of up to 0.72 (RMSE: 1.02 kcal/mol) on a single point. DynaMut2 predicts the variations in Gibbs Free Energy (ΔΔG) and in melting temperature (ΔTm) [8]. The dynamut2 analysis revealed that the missense mutation at the position 465 with replacement of Threonine to Isoleucine in the current predicted NAGPA 3D structure (Figure 1) will causes destabilization and the ΔΔG stability was brought down to -0.46 Kcal/mol. In the wild type, protein structure (Figure 2A) were Threonine at the position 465 displayed single hydrophobic interaction with Leucine at position 461 and several polar interactions with Leucine at 461, 462, Isoleucine at 463, Asparagine at 468 and leucine at 469. No direct contact was found with Alanine at 466 and 467. Ionic interactions were nil. On the contrary, in mutant type (Figure 2B) were Isoleucine present at the position 465 displayed two hydrophobic interactions with Leucine (462) and polar interactions are same as that of the wild type. Again, no ionic interactions were observed in the mutant type.
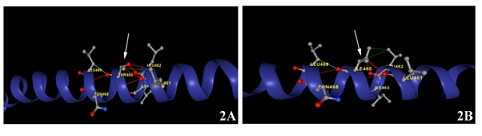 Figure 2: The dynamut2 analysis showingcomparison with wild type (Figure 2A) and the missense mutation at the amino acid position 465 with replacement of Threonine to Isoleucine in the variant type (Figure 2B) in the submitted NAGPA 3D structure. The amino acid change was highlighted with an arrow.
Figure 2: The dynamut2 analysis showingcomparison with wild type (Figure 2A) and the missense mutation at the amino acid position 465 with replacement of Threonine to Isoleucine in the variant type (Figure 2B) in the submitted NAGPA 3D structure. The amino acid change was highlighted with an arrow.
Mutagenic effect of rs7110: The RNA fold webserver output 3’UTR structure design is the secondary structure was based on the Minimum free energy (MFE) concept.The positions of loops and stems in both the structure (Figure 3A & 3B) are almost same except at the variant site (Figure 3B). The single nucleotide variation (SNV) is a change from residue “G” to “A” while “G” is a part of loop and “A” residue in the variant is a part of stem. There is a change in the position of loop and stem in the predicted secondary 3’UTR NAGPA structure.
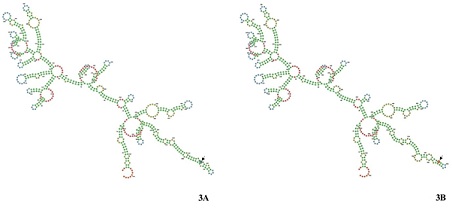 Figure 3: The image shows the RNA fold webserver output of secondary structure of 3’UTR region both wild type (Figure 3A) and variant type (Figure 3B). The single nucleotide variation (SNV) is a change from residue “G” to “A” while “G” is a part of loop and “A” residue in the variant is a part of stem was highlighted with an arrow.
Figure 3: The image shows the RNA fold webserver output of secondary structure of 3’UTR region both wild type (Figure 3A) and variant type (Figure 3B). The single nucleotide variation (SNV) is a change from residue “G” to “A” while “G” is a part of loop and “A” residue in the variant is a part of stem was highlighted with an arrow.
The 3’UTR mutation and miRNA site: The 3’UTR mutation most of the time creates a miRNA target site. In order to search for the miRNA target site that might have introduced due to c*527G>A(rs7110) variant, different miRNA scanning web servers were used. The scanning of 3’UTR region containing rs7110 mutation did not result any miRNA target site.
Discussion
According to some countable number of reports mutations in NAGPA gene were associated to non-syndromic stuttering [1,3]. Its almost 40 years since the first time NAGPA gene was reported [9,10] and about 20 years ago cloning was performed [11], The complete Human 3D structure of NAGPA gene yet to be published.Because of this constrain several questions related to NAGPA function, likethe mode of inhibition by its pro-peptide [12], its catalytic mechanism [13], and the role of its cysteine-rich C-Terminal Domain (CTD) [11] have remained unanswered. Few of the variants known to impair the nature of protein's folding and affect its activity [14], and some variants also found in control subjects [3]. Most of the variants reported so far lack the information about the effect on the enzyme activity and two such variants are rs7188856 and rs7110 as these two variants observed in our study we took this further to unravel its effect through In-silico based algorithms prediction.
A missense mutation may be deleterious as it formed due to alteration in the amino acid that usually produced at a specific site. Each amino acid has its own specific size, charge, and hydrophobicity value. The original wild-type residue and newly introduced mutant residue often differ in these properties. In the present study rs7188856 Threonine to Isoleucine at position 465 (T465I) a variant observed in NAGPA gene responsible for destabilising the structure. As reported elsewhere a threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia [15]. This needs to be further confirmed through biological assays. Generally, the variants found at the site of molecular interactions oftenalter binding energy and subsequently affects protein complex formation, on the contrary variants at the site of non-molecular interactions might reduce folding energy and it leads to monomer structure destabilisation [16].
The 3 UTR mutation rs7110 was earlier reported by Chen et al., [17]. It was observed during establishing an association between stuttering candidate genes GNPTAB, GNPTG and NAGPA with dyslexia in Chinese population [17]. The 3′ untranslated regions (3′ UTRs) of messenger RNAs (mRNAs) decides mRNA’s location, translation and its stability and also plays a major role in Protein–Protein Interactions (PPIs), and could able to transmit genetic information encoded in 3′ UTRs to proteins. This function has been shown to regulate diverse protein features, including protein complex formation or posttranslational modifications, but is also expected to alter protein conformations. Therefore, 3′ UTR-mediated information transfer can regulate protein features that are not encoded in the amino acid sequence [18]. Interestingly, differences in the 3′ UTRs can alter mRNAs localization near the synaptic region of neurons so that neuro transmitter synthesis can be regulated by synaptic demand and activity. As this can be correlated with the fact that the stuttering is a neurological disorder. Though final confirmation of variant-effect predictions has to come from experiments and computational methods will help in suggesting what experiments to do.
Conclusion
NAGPA mutations and stuttering, the connection between these words was established decade ago but it still demands strong evidence. There are hundreds of SNVs reported through GWAS analysis but major of these SNVs do not have supporting information regarding to their biological role. Such type study will help to select the more appropriate SNVs and they further can be taken for biological assays. In the current study, the two SNVs reported and their bioinformatic analysis suggests that these two variants can be further subjected to biological assays.
Acknowledgement
The authors would like to thank Dr. M Pushpavathi, Director and Dr. S R Savithri, former Director of All India Institute of Speech and Hearing, for granting permission to conduct the study. We also thank Dr. J S Jayashankar Rao and Dr. Srinivas Kovalli for their valuable support.
References
- Kang C (2013) Quantitative expression analysis of two NAGPA isoforms in multiple human cDNA tissue panels. Journal of Biomedical Research 14: 65-70.
- Kang C, Riazuddin S, Mundorff J, Krasnewich D, Friedman P, et al. (2010) Mutations in the lysosomal enzyme-targeting pathway and persistent stuttering. The New England journal of medicine 362: 677-685.
- Raza MH, Domingues CE, Webster R, Sainz E, Paris E, et al. (2016) Mucolipidosis types II and III and non-syndromic stuttering are associated with different variants in the same genes. European Journal of Human Genetics 24: 529-534.
- Gunasekaran N, Chandru J, Jeffrey JM, Mathuravalli K, Srisailapathy SRS, et al. (2021) Evaluation of recurrent GNPTAB, GNPTG, and NAGPA variants associated with stuttering. Advanced Genetics 2: 10043.
- Dusl M, Senderek J, Müller JS, Vogel JG, Pertl A, et al. (2015) A 3'-UTR mutation creates a microRNA target site in the GFPT1 gene of patients with congenital myasthenic syndrome. Human molecular genetics 24: 3418-3426.
- Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: 05005.
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, et al. (2003) MicroRNA targets in Drosophila. Genome Biol 5.
- Rodrigues CH, Pires DE, Ascher DB (2021) Dynamut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci 30: 60-69.
- Varki A, Kornfeld S (1980) Identification of a rat liver alpha-N-acetylglucosaminyl phosphodiesterase capable of removing "blocking" alpha-N-acetylglucosamine residues from phosphorylated high mannose oligosaccharides of lysosomal enzymes. J Biol Chem 255: 8398-8401.
- Waheed A, Hasilik A, Figura K (1981) Processing of the phosphorylated recognition marker in lysosomal enzymes. Characterization and partial purification of a microsomal alpha-N-acetylglucosaminyl phosphodiesterase. J Biol Chem 256: 5717-5721.
- Kornfeld R, Bao M, Brewer K, Noll C, Canfield W, et al. (1999) Molecular cloning and functional expression of two splice forms of human N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase. J Biol Chem 274: 32778-32785.
- Do H, Lee WS, Ghosh P, Hollowell T, Canfield W, et al. (2002) Human mannose 6-phosphate-uncovering enzyme is synthesized as a proenzyme that is activated by the endoproteasefurin. J Biol Chem 277: 29737-29744.
- Varki A, Sherman W, Kornfeld S (1983) Demonstration of the enzymatic mechanisms of alpha-N-acetyl-D-glucosamine-1-phosphodiester N-acetylglucosaminidase (formerly called alpha-N-acetylglucosaminylphosphodiesterase) and lysosomal alpha-N-acetylglucosaminidase. Arch Biochem Biophys 222: 145-149.
- Lee WS, Kang C, Drayna D, Kornfeld S (2011) Analysis of mannose 6-phosphate uncovering enzyme mutations associated with persistent stuttering. J Biol Chem 286: 39786-39793.
- Datta S, Quillin A, Rizig M, Blaveri E, Thirumalai S, et al. (2010) A threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia. Mol Psychiatry 5: 615-628.
- Teng S, Srivastava AK, Schwartz CE, Alexov E, Wang L, et al. (2010) Structural assessment of the effects of amino acid substitutions on protein stability and protein-protein interaction. Int J. of com. bio and drug design. 3: 334-349.
- Chen H, Xu J, Zhou Y, Gao Y, Wang G, et al. (2015) Association study of stuttering candidate genes GNPTAB, GNPTG and NAGPA with dyslexia in Chinese population. BMC Genetics 16: 7.
- Mayr C (2019) What Are 3' UTRs Doing? Cold Spring Harbor perspectives in biology 11: 034728.
Citation: Kundapur R, Sylvester C, Ramachandra NB, Maruthy S (2022) Predicting the mutagenic effects of variants rs7188856 and rs7110 of N-Acetylglucosamine-1-Phosphodiester Alpha-N-Acetylglucosaminidase (NAGPA) gene detected in randomly collected blood samples. J Genet Genomic Sci 7: 032.
Copyright: © 2022 Rajesh Kundapur, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

