
Regulatory Potential of Mobile Genetic Elements in the Human MGMT Gene
*Corresponding Author(s):
Lukash Lyubov LeonidovnaInstitute Of Molecular Biology And Genetics, National Academy Of Sciences Of Ukraine, 03680, 150 Zabolotnogo St., Kiev, Ukraine
Tel:+380 445260739,
Email:lukash.imbg@gmail.com
Abstract
Mobile Genetic Elements (MGE) make up a large part of the DNA of eukaryotes, in particular, almost 45% of the human genome. Numerous data show the diverse role of these elements in the genome from plasticity factors to mutability or stability. They discuss their role in the evolution of genomes and the evolution of gene regulation. The purpose of this work was to investigate the distribution of MGE in human MGMT gene and their regulatory potential. It has been shown at first that in the human MGMT gene, MGE is present both in the intron sequences and in the promoter region. In the intron sequences, MGEs form composite cluster structures that are the source of various regulatory sequences and have the potential to form alternative promoters. In the promoter region, three sequences of MGE were identified: two LTR repeats and a fragment of the DNA transposon. The MGE fragments in the promoter region of human MGMT also enriched with potential cis-regulatory sequences that may be involved in the regulation of this gene.
Keywords
INTRODUCTION
The eukaryotic genome is a complex and dynamic structure. About 50% of the human genome, and possibly more is covered by Mobile Genetic Elements (MGE), which for a long time were considered unnecessary ballast [1,2]. Studies of recent years convincingly testify to the important role of these elements in the evolution of genomes and in the evolution of gene regulation [3-15]. They are not only mutagenic factors, but they can also be the source of a variety of regulatory sequences, such as sites of alternative splicing, cis-regulatory modules, which are clusters of binding sites of transcription factors, or play the role of alternative promoters [16-24]. MGE tends to integrate into non-coding genome sites (introns, flanked genes and intergenic sites) [25]. On average, in the human introns, MGE is approximately 89.5%; in exons, they account for slightly more than 10%, in particular about 4% for protein-encoding genes [26]. Up to 20% of the genes contain MGE in non-translated regions of mRNA, where they can affect the regulation of gene expressions; in particular, the MGE in 5'UTR affects the initiation of translation [27]. It is known that about 25% of human genes contain MGE in promoter regions, and today there is convincing evidence of their involvement, in particular promoters of retroelements, in the regulation of gene transcriptional activity [21,26]. There are cases where MGE plays the role of alternative promoters, which leads either to increase the level of expression of the corresponding gene, or to change the tissue specificity of its expression [28].
Human MGMT gene encodes a reparative enzyme called O6-methylguanine-DNA methyltransferase, which removes alkylated groups from the O6 position of guanine in DNA and protects cells from their toxic and mutagenic effects. Expression of this gene and the activity of the enzyme themselves have wide limits both in between and intrinsically individual variations, indicating that its regulation is complicated [29]. Given the complexity of this problem, it might be worthwhile to focus on MGE, the role of which in gene regulation has recently been discussed quite widely. Numerous databases today have enormous material on the availability of mobile elements, but the data array is not characterized and not generalized. Perhaps the MGE of MGMT gene is also a source of a variety of regulatory sequences, which led us to focus our attention on the study of this issue.
MATERIALS AND METHODS
The nucleotide sequence of the MGMT gene is taken on the Ensembl site. Data on the promoter region of the gene were obtained from GenBank (X61657), about the potential promotor regions of the gene being studied from the AceView database. The results of the search and identification of IHE are done with the CENSOR program. The homology between the sequences studied was determined by the BLAST 2.2.32 program. Functional sites are defined by TFSEARCH: Searching Transcription Factor Binding Sites (ver 1.3). The search for potential regulatory sequences was performed by SITECON, SiteGA, BLASTN 2.2.26 using the TRRD database resources (Transcription Regulatory Regions Database) and CISTER: Cis-element Cluster Finder, WWW Signal Scan and Tfsitescan. These methods and databases were used in the relevant sections of the article.
RESULTS AND DISCUSSION
Distribution of MGE in the gene of human MGMT reparative enzyme
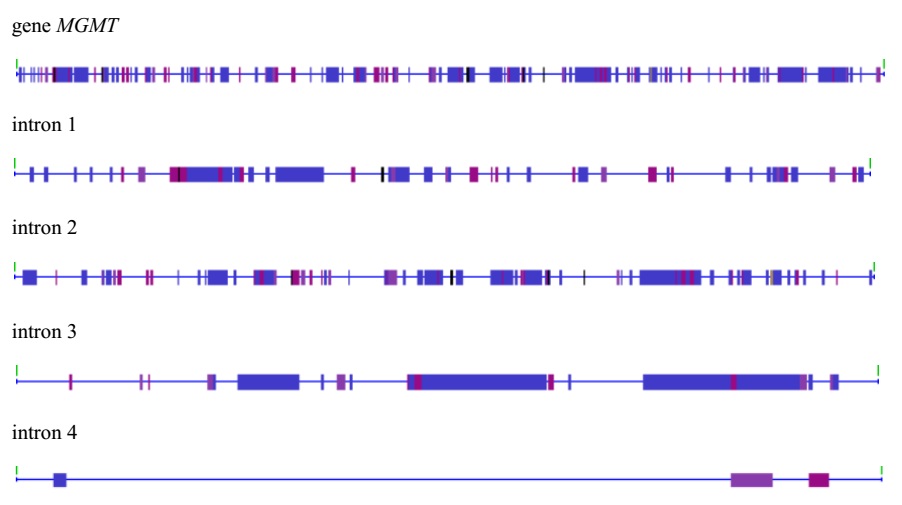
Figure 1: MGE in the human MGMT gene and in its introns.
| Intron number (bp) | % MGE, (bp) | ||||
| DNA- transposons | Endogenous retroviruses | LTR retrotransposons | Non-LTR retrotransposons | In total | |
| intron 1 (68.944) | 6.05 (4169) | 2.36 (1625) | 19.26 (13281) | 27.67 (19075) | |
| intron 2 (171.517) | 4.14 (7102) | 2.19 (3762) | 0.24 (409) | 20.75 (35590) | 27.32 (46863) |
| intron 3 (51.158) | 2.26 (1158) | 2.63 (1345) | 39.85 (20388) | 44.74 (22891) | |
| intron 4 (7.446) | 2.25 (167) | 4.71 (351) | 1.3 (97) | 8.26 (615) | |
| Gene (300824) | 4.21 (12676) | 2.35 (7083) | 0.24 (409) | 23.03 (69275) | 29.73 (89443) |
| Coordinates within the intron | MGE | Length bp | Class / family | Chain | Number cluster |
| 103739-104018 | AluSg | 280 | NonLTR/SINE/SINE1 | - | 1 |
| 104019-104712 | L1ME_ORF2 | 694 | NonLTR/L1 | + | 1 |
| 104713-105077 | L1ME_ORF2 | 365 | NonLTR/L1 | + | 1 |
| 125707-125739 | MuDRF-2_MLP | 33 | DNA/MuDR | + | 2 |
| 125742-125938 | L1MC4_5end | 211 | NonLTR/L1 | + | 2 |
| 125941-126057 | MER81 | 115 | DNA/hAT | - | 2 |
| 126058-126151 | L1MC4_5end | 101 | NonLTR/L1 | + | 2 |
| 150912-151320 | MER61E | 414 | LTR | - | 3 |
| 151322-152319 | L1HS | 1005 | NonLTR/L1 | - | 3 |
| 152320-152398 | L1-2_Cja | 79 | NonLTR/L1 | + |
3 |
Table 2: MGE clusters in human MGMT intron 2.
| Coordinates within the intron | MGE | Length bp | Class / family | Chain | Number cluster |
| 13431-14056 | L1MEC_5 | 626 | NonLTR/L1 | + | 1 |
| 14060-14677 | L1ME_ORF2 | 618 | NonLTR/L1 | + | 1 |
| 14681-14963 | AluSz6 | 283 | NonLTR/SINE/SINE1 | + | 1 |
| 24109-24489 | L1PA6 | 378 | NonLTR/L1 | - | 2 |
| 24490-30537 | L1HS | 6049 | NonLTR/L1 | - | 2 |
| 30538-30654 | MER81 | 120 | NonLTR/L1 | - | 2 |
| 30655-31384 | L1-2_Cja | 733 | NonLTR/L1 | + | 2 |
| 42743-42794 | L1MC1_EC | 52 | NonLTR/L1 | - | 3 |
| 42795-45806 | L1MB3_EC | 3223 | NonLTR/L1 | - | 3 |
| 45807-45933 | L1MEf_5end | 140 | NonLTR/L1 | - | 3 |
| 45941-46419 | L1MD1_5 | 476 | NonLTR/L1 | - | 3 |
Table 3: MGE clusters in human MGMT intron 3.
Composite cluster structures of MGE in the introns of the investigated gene as a source of potential regulatory sequences
|
Trans-cription factor, |
Ingredients of composite clusters | |||||
| intron 2 (cluster 1) | intron 3 (cluster 1) | |||||
| AluSg | L1ME_ORF2 | L1ME_ORF2 | L1MEC_5 | L1ME_ORF2 | AluSz6 | |
| C/EBP | + | + | + | + | + | |
| 1-Oct | + | + | + | + | + | |
| SRY | + | + | + | + | + | |
| GATA | + | + | + | + | + | |
| HSF2 | + | + | + | + | ||
| AP-1 | + | + | + | |||
| Pbx-1 | + | + | + | |||
| MZF1 | + | + | + | |||
| TATA | + | + | ||||
| STAT | + | + | ||||
| RORalp | + | + | ||||
Table 4: Representation of potential sites of binding the boundaries of composite cluster structures of MGE in human MGMT gene introns.
Motives of regulatory sequences in the promoter regions of the MGMT gene within the MGE
The promoted gene promoter (X61657) has a length of 1157 bp and covers exon 1 part intron 1. It is devoid of TATA or CAAT sequences, contains CG-rich regions and is structurally reminiscent of genes of the household. It also contains SP1, AP-1 and AP-2, NF-kapBsites, two elements that bind the Glucocorticoid Receptor (GRE) and a 59-bp size element, which is located on the first eccentric-intron boundary, necessary for effective transcription of the reporter designs [46,47].
Fragments of MGE in the promoter region of the investigated gene
| Referential promoter, symbolic | Data on MGE | ||||
| Element | Class | Length | Direction | Coordinates within promotor | |
| MLT1C2 | ERV/ERV3 | 117 | c | -949/-839 | |
| hO6P | MLT1C | ERV/ERV3 | 147 | c | -813/-667 |
| SETARIA1 | DNA | 341 | D | -265/+76 | |
Table 5: Fragments of MGE in the promoter region of human MGMT gene.
Potential cis-elements within the fragments of mobile genetic elements in the promoter regions of the investigated gene
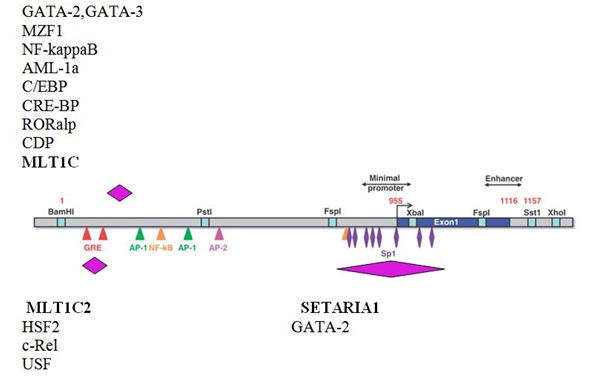
Figure 2: Potential cis-regulatory elements in the MGE fragments identified within the sequence of the refracted promoter of human MGMT gene.
Fragments of the MGE as components of potential alternative promoters
Fragments of the MGE in alternative promoter regions of the investigated gene
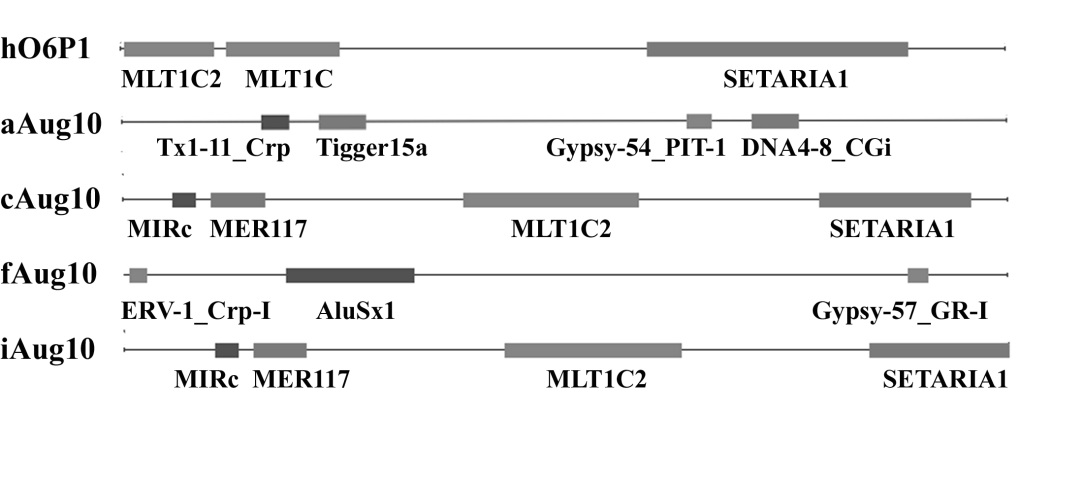
| Symbolic designation | Data on MGE | ||||
| for the promoter | Element | Class | Length | Direction | Coordinates within promoter |
| aAug10 | Tx1-11_Crp | NonLTR/Tx1 | 60 | c | 320-379 |
| Tigger15a | DNA/Mariner | 105 | c | 448-552 | |
| Gypsy-54_PIT-I | LTR/Gypsy | 54 | d | 1278-1331 | |
| DNA4-8_CGi | DNA | 104 | d | 1425-1528 | |
| cAug10 | MIRc | NonLTR/SINE/SINE2 | 53 | c | 115-167 |
| MER117 | DNA/hAT | 120 | d | 202-321 | |
| MLT1C2 | ERV/ERV3 | 397 | c | 771-1167 | |
| SETARIA1 | DNA | 341 | d | 1576-1916 | |
| fAug10 | ERV-1_Crp-I | ERV | 38 | c | 19-56 |
| AluSx1 | NonLTR/SINE/SINE1 | 285 | c | 374-658 | |
| Gypsy-57_GR-I | LTR/Gypsy | 44 | c | 1777-1820 | |
| iAug10 | MIRc | NonLTR/SINE/SINE2 | 53 | c | 208-260 |
| MER117 | DNA/hAT | 120 | d | 295-414 | |
| MLT1C2 | ERV/ERV3 | 397 | c | 864-1260 | |
| SETARIA1 | DNA | 314 | d | 1685-1998 | |
Table 6: Sequence of MGE in potential alternative promoter regions of the MGMT human reparative enzyme gene.
Potential regulatory sequences within identified MGE fragments
| Conditional designation of the promoter | Mobile genetic elements | Regulatory sequences | ||
| Name | Species affiliation | Linking sites for transcription factors | Regulatory elements | |
|
aAug10 fAug10 |
Tigger15a AluSx1 |
Mammalia Primates |
ERE; C/EBP TATA box; YY1; GATA; SF1;Sp1;CAAT box; OCT1; SREBP; PAX2;
|
?romoter; Locus Control Region-like region promoter;
|
Table 7: Regulatory potential sequences of MGE, inherent in the genome of the man, within the potential promoters of the human MGMT gene.
CONCLUSION
In the human MGMT gene, MGE is present both in the intron sequences and in the promoter region. In the intron sequences, MGEs form composite cluster structures that are the source of various regulatory sequences and have the potential to form alternative promoters. In the promoter region, three sequences of MGE were identified: two LTR-repeats and a fragment of the DNA-transposon. The fact that one of the LTR-repeats contains the described response elements of the Glucocorticoid hormones (GRE) and that the minimal promoter and sequence of the SP1 site are located within the fragment of DNA-transposon confirms the important role of MGE in gene regulation. In addition, the MGE fragments in the promoter region of human MGMT enriched with potential cis regulatory sequences that may also be involved in the regulation of this gene.
REFERENCES
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860-921.
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD (2011) Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: 1002384.
- Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr (2003) Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 13: 651-658.
- Hedges DJ, Batzer MA (2005) From the margins of the genome: mobile elements shape primate evolution. Bioessays 27: 785-794.
- Werren JH (2011) Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci USA 108: 10863-10870.
- Piskurek O, Jackson DJ (2012) Transposable elements: From DNA parasites to architects of metazoan evolution. Genes (Basel) 3: 409-422.
- Chalopin D, Naville M, Plard F, Galiana D, Volff JN (2015) Comparative analysis of Transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol 7: 567-580.
- Platt RN, Vandewege MW, Ray DA (2018) Mammalian transposable elements and their impacts on genome evolution. Chromosome Res 26: 25-43.
- Medstrand P, vande Lagemaat LN, Dunn CA, Landry JR, Svenback D, et al. (2005) Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet Genome Res 110: 342-352.
- Feschotte C (2008) Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397-405.
- Jurka J (2008) Conserved eukaryotic transposable elements and the evolution of gene regulation. Cell Mol Life Sci 65: 201-204.
- Bourque G (2009) Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev 19: 607-612.
- Rayan NA, Del Rosario RCH, Prabhakar S (2016) Massive contribution of transposable elements to mammalian regulatory sequences. Semin Cell Dev Biol 57: 51-56.
- Chuong EB, Elde NC, Feschotte C (2017) Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71-86.
- Sundaram V, Wang T (2018) Transposable Element Mediated Innovation in Gene Regulatory Landscapes of Cells: Re-Visiting the "Gene-Battery" Model. Bioessays 40.
- Callinan PA, Batzer MA (2006) Retrotransposable elements and human disease. Genome Dyn 1: 104-115.
- Chenais B (2015) Transposable elements in cancer and other human diseases. Curr Cancer Drug Targets 15: 227-242.
- Kazazian HH Jr, Moran JV (2017) Mobile DNA in Health and Disease. N Engl J Med 377: 361-370.
- Sorek R, Ast G, Graur D (2002) Alu-Containing Exons are Alternatively Spliced. Genome Res 12: 1060-1067.
- Stower H (2013) Alternative splicing: Regulating Alu element 'exonization'. Nat Rev Genet 14: 152-153.
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV (2003) Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet 19: 68-72.
- Thornburg BG, Gotea V, Maka?owski W (2006) Transposable elements as a significant source of transcription regulating signals. Gene 365: 104-110.
- Rebollo R, Romanish MT, Mager DL (2012) Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21-42.
- Batut P, Dobin A, Plessy C, Carninci P, Gingeras TR (2013) High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res 23: 169-180.
- Prak ET, Kazazian HH Jr (2000) Mobile elements and the human genome. Nat Rev Genet 1: 134-144.
- Nekrutenko A, Li WH (2001) Transposable elements are found in a large number of human protein-coding genes. Trends Genet 17: 619-621.
- van de Lagemaat LN, Landry JR, Mager DL, Medstrand P (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19: 530-536.
- Faulkner GJ, Carninci P (2009) Altruistic functions for selfish DNA. Cell Cycle 8: 2895-2900.
- Christmann M, Verbeek B, Roos WP, Kaina B (2011) O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: enzyme activity, promoter methylation and immunohistochemistry. Biochim Biophys Acta 1816: 179-190.
- Jurka J, Kohany O, Pavlicek A, Kapitonov VV, Jurka MV (2004) Duplication, coclustering, and selection of human Alu retrotransposons. Proc Natl Acad Sci USA 101: 1268-1272.
- Kolomietz E, Meyn MS, Pandita A, Squire JA (2002) The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer 35: 97-112.
- Kaer K, Branovets J, Hallikma A, Nigumann P, Speek M (2011) Intronic L1 retrotransposons and nested genes cause transcriptional interference by inducing intron retention, exonization and cryptic polyadenylation. PLoS One 6: 26099.
- Kaer K, Speek M (2012) Intronic retroelements: Not just “speed bumps” for RNA polymerase II. Mob Genet Elements 2: 154-157.
- Wheelan SJ, Aizawa Y, Han JS, Boeke JD (2005) Gene-breaking: A new paradigm for human retrotransposon-mediated gene evolution. Genome Res 15: 1073-1078.
- Oei SL, Babich VS, Kazakov VI, Usmanova NM, Kropotov AV, et al. (2004) Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters. Genomics 83: 873-882.
- Thomas MJ, Seto E (1999) Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236: 197-208.
- Yang WM, Inouye C, Zeng Y, Bearss D, Seto E (1996) Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA 93: 12845-12850.
- Oei SL, Griesenbeck J, Schweiger M, Babich V, Kropotov A, et al. (1997) Interaction of the transcription factor YY1 with human poly(ADP-ribosyl) transferase. Biochem Biophys Res Commun 240: 108-111.
- Griesenbeck J, Ziegler M, Tomilin N, Schweiger M, Oei SL (1999) Stimulation of the catalytic activity of poly(ADP-ribosyl) transferase by transcription factor Yin Yang 1. FEBS Lett 443: 20-24.
- Kakizawa T, Miyamoto T, Ichikawa K, Kaneko A, Suzuki S, et al. (1999) Functional interaction between Oct-1 and retinoid X receptor. J Biol Chem 274: 19103-19108.
- Umesono K, Murakami KK, Thompson CC, Evans RM (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65: 1255-1266.
- Mangelsdorf DJ, Evans RM (1995) The RXR heterodimers and orphan receptors. Cell 83: 841-850.
- Speek M (2001) Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 21: 1973-1985.
- Matlik K, Redik K, Speek M (2006) L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol 2006: 71753.
- Wheelan SJ, Aizawa Y, Han JS, Boeke JD (2005) Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res 15: 1073-1078.
- Harris LC, Potter PM, Tano K, Shiota S, Mitra S, et al. (1991) Characterization of the promoter region of the human O6-methylguanine-DNA methyltransferase gene. Nucleic Acids Res 19: 6163-6167.
- Harris LC, Remack JS, Brent TP (1994) Identification of a 59 bp enhancer located at the first exon/intron boundary of the human O6-methylguanine DNA methyltransferase gene. Nucleic Acids Res 22: 4614-4619.
- Polak P, Domany E (2006) Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics 7: 133.
- Usmanova NM, Kazakov VI, Tomilin NV (2008) [Retroposons of Alu family from cis-regulatory modules of DNAse II and CAML genes effect gene expression in A549 and HEK293 cells]. Tsitologiia 50: 249-255.
- Bannert N, Kurth R (2004) Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA 101: 14572-14579.
- Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL (2007) Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet 3: 10.
- Sverdlov ED (1998) Perpetually mobile footprints of ancient infections in human genome. FEBS Lett 428: 1-6.
- Shankar R, Grover D, Brahmachari SK, Mukerji M (2004) Evolution and distribution of RNA polymerase II regulatory sites from RNA polymerase III dependant mobile Alu elements. BMC Evol Biol 4: 37.
- Vansant G, Reynolds WF (1995) The consensus sequence of a major Alu subfamily contains a functional retinoic acid response element. Proc Natl Acad Sci USA 92: 8229-8233.
- Laperriere D, Wang TT, White JH, Mader S (2007) Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics 8: 23.
- Babich V, Aksenov N, Alexeenko V, Oei SL, Buchlow G, et al. (1999) Association of some potential hormone response elements in human genes with the Alu family repeats. Gene 239: 341-349.
- Piedrafita FJ, Molander RB, Vansant G, Orlova EA, Pfahl M, et al. (1996) An Alu element in the myeloperoxidase promoter contains a composite SP1-thyroid hormone-retinoic acid response element. J Biol Chem 271: 14412-14420.
- Hamdi HK, Nishio H, Tavis J, Zielinski R, Dugaiczyk A (2000) Alu-mediated phylogenetic novelties in gene regulation and development. J Mol Biol 299: 931-939.
- Romanish MT, Nakamura H, Lai CB, Wang Y, Mager DL (2009) A Novel Protein Isoform of the Multicopy Human NAIP Gene Derives from Intragenic Alu SINE Promoters. PLoSOne 4: 5761.
- Bolotin E, Chellappa K, Hwang-Verslues W, Schnabl JM, Yang C, et al. (2011) Nuclear receptor HNF4α binding sequences are widespread in Alu repeats. BMC Genomics 15: 560.
- 61. Pidpala OV, Lukash LL (2015) Motives of regulatory sequences in human promotor regions of human MGMT within the framework of mobile genetic elements. Factors of experimental evolution of organisms 16.
- Mayorov VI, Rogozin IB, Elisaphenko EA, Adkison LR (2000) B2 elements present in the human genome. Mamm Genome 11: 177-179.
- Pidpala OV, Yatsishina AP, Lukash LL (2006) Mobile genetic elements in the genome-oncosuppressor p53: evolutionary aspect. Factors In Experimental Evolution of Organisms 3: 43-48.
- Pidpala OV, Yatsishina AP, Lukash LL (2007) Fragments of bacterial IS-elements and mobile genetic elements of eukaryotes in human mt DNA. Achievements and problems genetics, breeding and biotechnology 1: 498-502.
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, et al. (2009) The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563-571.
- de Souza FS, Franchini LF, Rubinstein M (2013) Exaptation of transposable elements into novel cis-regulatory elements: is the evidence always strong? Mol Biol Evol 30: 1239-1251.
- Patrushev LI, Kovalenko TF (2014) Functions of noncoding sequences in mammalian genomes. Biochemistry (Mosc) 79: 1442-1469.
Citation: Pidpala O, Leonidovna LL (2018) Regulatory Potential of Mobile Genetic Elements in the Human MGMT Gene. J Genet Genomic Sci 3: 008.
Copyright: © 2018 Lukash Lyubov Leonidovna, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

