
Relationship between Skin Microbiota and Skin Biophysical Parameters in Inflammatory Skin Disease: A Systematic Review
*Corresponding Author(s):
Paola PeruginiDepartment Of Drug Sciences, Via Taramelli 12, University Of Pavia, 27100 Pavia, Italy
Tel:+39 0382987174,
Email:paola.perugini@unipv.it
Abstract
Many recent studies highlight the importance of skin microbiota for skin health. Alterations in the balance of bacterial flora cause the development of inflammatory skin diseases such as acne, atopic dermatitis, or psoriasis.
This systematic review aims to investigate the relationship, in these skin diseases, between skin microbiota and skin biophysical parameters, such as pH, Transepidermal Water Loss (TEWL), Hydration (HI) and sebum levels.
Google Scholar, Medline via Pubmed, and Web of Science were considered as scientific database to search studies about this topic. Research about acne and psoriasis did not produce any results. For this reason, in this review, only articles concerning atopic dermatitis were discussed. Therefore, a possible correlation between skin barrier functionality and microbial composition was also investigated. So, this could be a starting point for the diagnosis of atopic dermatitis or, more generally, for all inflammatory skin diseases.
Introduction
Human skin is an interface between the human body and the external environment, and it plays an important role as a chemical and physical barrier.
The skin is covered with many microorganisms (about one million per square centimeter) such as viruses, bacteria, and fungi. All these microorganisms are defined as skin microbiota: it is characteristic and specific for everyone. Between these microbial communities and skin is established a symbiotic relationship. It is essential for maintaining an adequate balance, and therefore healthy skin [1-4].
The alteration of this balance takes the name of dysbiosis and determines the onset of skin diseases, such as acne, atopic dermatitis, and psoriasis. These are associated with imbalances of the skin microbiota, in which the abundance and diversity of bacterial species changes [4-6].
Atopic dermatitis (AD) is a chronic inflammatory skin disease in which a key role seems to be played by Staphylococcus aureus. In fact, in atopic dermatitis, patients are more likely to be colonized by S. Aureus than in healthy controls. Moreover, the enhancement of colonization of S. aureus increases the severity of dermatitis [7]. S. aureus determines long-lasting local inflammation and local immunosuppression by induction of suppressors cells, binding the Toll-Like Receptor (TRL2) [8,9].
A major risk factor for the onset of atopic dermatitis is a mutation in the FLG gene. The FLG gene encodes for the filaggrin protein, which plays a key role in skin barrier function. It maintains the structural and mechanical integrity of the stratum corneum and guarantees homeostasis of the epidermal barrier. Consequently, mutations or deficiencies of FLG determine an anomaly in the permeability of the barrier function. This disease is characterized by an erythematous skin rash that can progress to redness and scaling. Also, xerosis cause rough, scaly, or cracked skin [10-12].
Psoriasis is a skin disease characterized by epidermal hyperproliferation, inflammation of the dermis, and hyperkeratosis with the subsequent formation of thickened and scaly plaques.
To date, the exact cause of its onset is still unknown. However, it is supposed that there is an inappropriate activation of the skin's immune system against pathogens [3,13,14].
In psoriasis, the role played by superantigens is very important. These are viral or bacterial proteins that stimulate the proliferation of T-lymphocytes in the skin. In this way, the inflammatory response is triggered. Although it seems well established that streptococcal infection is significant in psoriasis, it appears that other microorganisms may also be involved in the pathogenesis of this disease [15-17].
In fact, in exacerbations of psoriasis, also S. aureus has been identified. S. aureus activates Th1 and Th17 cells, promoting the production of interleukins. They stimulate keratinocytes to produce chemokines, cytokines, and proinflammatory mediators, thus leading to chronic inflammation [18,19].
Acne vulgaris is a chronic inflammatory disease characterized by an increase in sebum production, alteration of the quality of sebum lipids, follicular hyperkeratinisation, and inflammation. In all this, a key role is played by Propionibacterium acnes, who mainly colonized the sebaceous areas and hydrolysing sebum’s triglycerides, producing free fatty acids, which acidify and weaken the skin [3,20,21]. P. acnes secretes lipases, metalloproteases, and porphyrins. These react with molecular oxygen, generating toxic species that cause damage to keratinocytes.
Moreover, P. acnes interacts with markers of innate immunity such as TLR and AMP. The prolonged activation of innate immunity, maintained by cytokine production, plays an important role in the chronic evolution of acne lesions [22-24].
Therefore, all these chronic inflammatory diseases manifest themselves with alterations in the appearance of the skin. Consequently, these changes cause variations in skin biophysical parameters, such as Hydration, TEWL, roughness, and corneocytes’ desquamation. So, this review aims to investigate in the literature the correlation between the presence of specific microorganisms and skin biophysical parameters in this disease.
Materials And Methods
Google Scholar, Medline via Pubmed, and Web of Science were systematically searched for studies about the relationship between skin parameters, such as pH, Transepidermal Water Loss (TEWL), Hydration (HI), sebum levels, and microorganisms in skin diseases. Specifically, the research focused on these diseases: atopic dermatitis, psoriasis, and acne. Also, to identify further considerable studies was carried out a reference check.
Experimental and observational studies performed on patients of all ages were included. Case reports were excluded.
Results were screened up to January 7, 2021. No restrictions were applied about language and publication date.
Initially, titles and abstracts were screened for relevance. Articles containing in general analyses of skin microbiota and skin biophysical parameters were selected. Afterwards, full texts were read and screened according to the inclusion criteria: studies concerning microbiota analysis in patients with atopic dermatitis, acne, psoriasis, and studies concerning biophysical skin parameters in these patients were included.
After a process of article screening and selection, only papers including analysis of both skin microbiota and biophysical skin parameters in patients with atopic dermatitis, acne, and psoriasis were considered.
The included studies were summarized in the Table 1. Then, they were critically discussed.
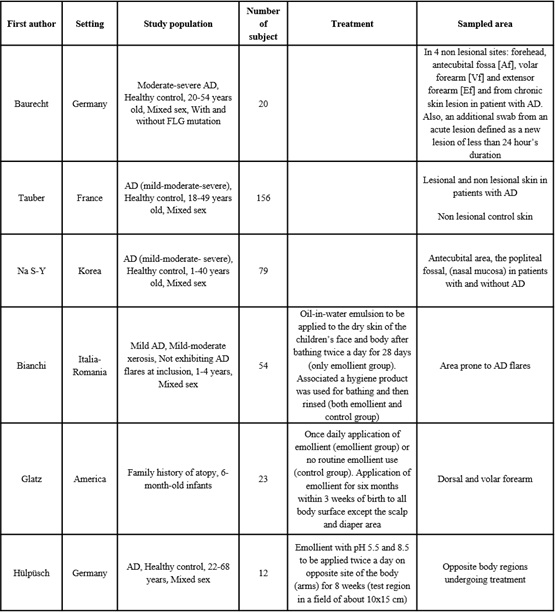 Table 1: Main parameters of all studies included in this review.
Table 1: Main parameters of all studies included in this review.
Results
Figure 1 reports the flowchart describing the paper screening process and their selection strategy. During database searching, 2953 studies were identified. Based on the title and abstract, 247 papers were screened. Fifty-four records met our inclusion criteria after reading the full text: 31 about atopic dermatitis, 12 on psoriasis, and 15 on acne. Finally, only six records were included in this analysis, all about atopic dermatitis; no study in patients with psoriasis or acne met our final criteria.
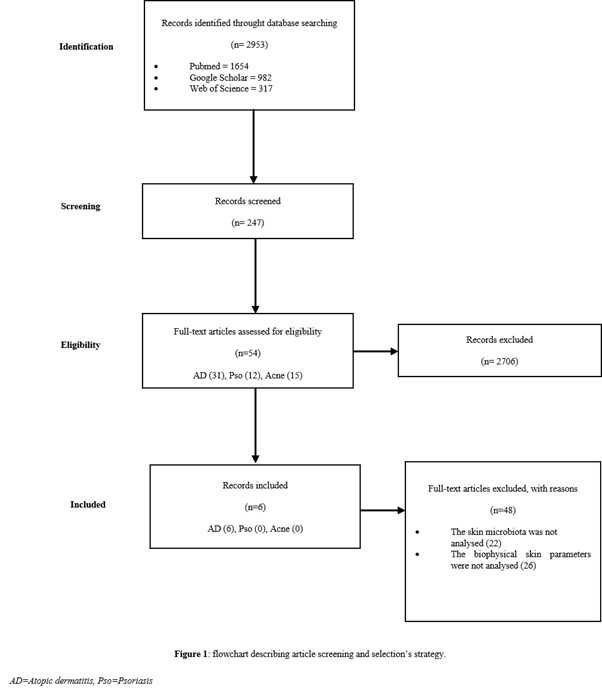 Figure1: Flowchart describing article screening and selection’s strategy.
Figure1: Flowchart describing article screening and selection’s strategy.
Discussion
Description of the study
Six observational studies were included in this systematic review, all about atopic dermatitis. In fact, research about acne and psoriasis did not produce any results. These six studies, including 344 patients with AD, were investigated to identify a correlation between alterations in skin microbiota and variation of skin biophysical parameters. The ages of the participants ranged from 6 months to 54 years, and subjects of both genders were included. In all studies, the severity of the disease was defined by the authors using the SCORAD index [25-30].
The first study analysed comprehensively the microbiome of 10 patients with AD and 10 healthy subjects, matched by age and sex. In these two groups, there were 5 patients with FLG mutation and 5 without FLG mutation. Analyses were conducted in four different sites (forehead, antecubital fossa, volar forearm, and extensor forearm). Furthermore, in AD subjects, analyses were also performed on the acute and chronic lesions. On the same sampling sites, skin’s physiology parameters, including lipid profile, were measured to explore the correlation between skin microbiota and epidermal barrier function abnormalities in patients with AD [25].
In the second and third articles, the relationship between the abundance of S. aureus and the skin's barrier function was evaluated. These studies were conducted comparing two groups of subjects: a group of patients with AD and a control group made up of healthy subjects [26,27].
In Tauber, et al. 78 patients with AD were matched to a control group, also made up of 78 subjects [26]. While in Na S-Y, et al. 39 patients with AD were enrolled and compared with 40 healthy controls [27]. Patients and controls were matched for age and gender. All analyses were performed on lesion areas in AD patients and in the same skin location of matched controls [26,27].
In the last three studies, the analysis of the skin microbiota and barrier function parameters was carried out in function of treatment, dividing the subject randomly into two groups. The primary objective was to evaluate the effect of an emollient treatment. However, were also analysed the skin microbiota and the skin physiological parameters. So, attention was also paid to these aspects [28-30].
In Pascale, et al. 54 children (1-4 years old) with AD were randomized into two groups, the emollient one intended to receive the treatment, and the control one [28]. In Martin, et al. the 24 subjects, enrolled to be randomized in two groups, were 6-month-old children with a family history of atopy such as dermatitis or asthma [29]. Finally, in Hülpüsh, et al. unlike the two previous studies, in addition to patients with AD, there were also healthy control groups [30].
In summary, in the first three studies considered, skin microbiota and skin physiological parameters, in patients with AD and healthy subjects, were compared [25-27].
While in the other three studies an emollient treatment was used to evaluate the progress from the point of view of the skin microbiota and from that of physical barrier parameters [28-30].
In two studies, the measurements were made at T0 and at the end of the treatment, so there is a baseline value [28,30]. While in the study conducted by Martin, et al. the analyses were conducted only after six months of using the product or not. In this case, the lack of basal values represents a significant limit [29]. Also, only in one study there is a healthy control group, formed by subjects without AD [30].
However, in all studies the number of subjects enrolled is a little small. In some cases homogeneous, in others heterogeneous from the point of view of age: this is a notable limitation.
Methodology of analysis
To carry out the analysis of the microbiota the skin was sampled mainly using swabs. However, different protocols were used for DNA extraction. In four studies, bacterial DNA was amplified, and later the 16S rRNA gene was sequenced, targeting the hypervariable region V1-V3. In these studies, were reported percentages on bacterial communities at the level of genus, family, and species [25,28-30].
In two studies were not analysed the entire microbiota, but was conducted only the Staphylococci research. In these cases, the presence of Staphylococcus aureus was confirmed by a coagulase test [26,27].
Regarding skin parameters, for evaluation of barrier function, skin hydration (HI) was measured in three studies [26,29,30]. The pH of the skin was measured in four studies [25,26,29,30]. Finally, the Transepidermal Water Loss (TEWL) was measured in all studies [25-30]. Also, Baurecht, et al. analysed the skin lipid composition using a mass spectrometer [25].
Profile of skin microbiota in atopic dermatitis
As previously clarified, alterations in the balance of skin microbiota cause the onset of chronic inflammatory diseases. In fact, from all studies, it emerged that in subjects with AD there is a distinct microbial configuration compared to healthy subjects. As reported, by Baurecht et al. even in sites not strictly affected by AD, the microbial composition appears to be different compared with the one of a healthy individual [25].
Moreover, sites affected by AD showed a decrease in the diversity and richness of many genres. In subjects with AD, compared to healthy controls, significantly increase the amount of S. aureus, especially in acute and chronic lesions. Also, the abundance of this bacterium was correlated positively with the severity of the disease. The severity of the disease was measured with the SCORAD index [25-30]. This concept emerged clearly from studies that involve the use of a treatment, in which the abundance of S. aureus increased significantly in untreated patients [27,28].
In the study conducted by Na S-Y, et al. S. aureus was also isolated from the nasal mucosa, suggesting the hypothesis that the nose may be an important reservoir of S. aureus strains, that can spread on the skin [27].
Also, it is interesting to underline the role of the FLG gene: a gene that codes for a protein that, as mentioned, causes the impairment of the skin barrier by reducing the integrity of the stratum corneum. Also, it affects the formation of lipids and increases the growth of S. aureus [31].
In Baurecht, et al. healthy subjects with FLG mutation shown a consistent decrease in bacterial diversity and richness, besides an imbalance between some species and an increase in the Staphylococci genus. This is the reason why the skin microbiota composition of healthy subjects FGL deficient is more similar to the one of AD patients than to the one of healthy subjects [25].
However, in some of these studies, emerges an important limitation: the low prevalence of S. aureus in xerotic areas. This is mainly due to the mild severity of the AD of the subject involved [28,30].
Skin physiological parameters in atopic dermatitis
The skin barrier plays a key role in the clinical expression of AD. The evaluation of skin physiological parameters can be predictive of the functionality of the skin barrier. Among the various parameters, the maintenance of an acid skin pH is essential for a functional skin barrier. An increase in pH can compromise the integrity of the stratum corneum and of skin barrier function. Thus, altered skin pH may contribute to the breakdown of the skin barrier seen in AD patients [32,33]. A breakdown of the skin barriers is associated with a decrease in skin hydration and an increase in TEWL [34].
In all the studies considered, the skin of AD patients, when compared with control subjects, presents anomalies in the skin barrier, and so altered physical and chemical properties.
In fact, the skin of AD patients shows higher pH and TEWL and lower hydration degree than healthy controls. But most of all, the increase in TEWL and the decrease in hydration are statistically correlated with the clinical severity of the disease [26-28,30].
Effects of treatment on the skin microbiota and skin physiological parameters in atopic dermatitis
Three of the studies considered involved the use of an emollient treatment. In all these studies, the daily use of an emollient was able to improve mild AD. The treatment appears to protect the skin from an increase in S. aureus. In fact, the levels of this bacterium increased in the control group but remained unchanged in the emollient group [28]. More generally, in two other studies, in which microbial diversity and richness were analysed, it has been confirmed that the emollient treatment can maintain greater bacterial diversity [29,30].
Moreover, the treatment with emollient seems to restore the functionality of the skin barrier. It has been proved by a reduction in TEWL and maintenance of a lower pH [28-30].
The main limitation of these studies is that the treatment with emollient lasted for a short period, so it is not possible to assert if the emollient can really postpone AD flare by maintaining an adequate balance of cutaneous microflora and correct barrier functionality.
Conclusion
As previously clarified, the clinical expression of some chronic inflammatory skin diseases, such as acne, atopic dermatitis, and psoriasis, is the result of dysbiosis conditions of the skin microbiota and alterations in the functionality of the skin barrier. This systematic review aims to investigate, in literature, a possible correlation between these two aspects, so that they can be used to support the other. The analysis focused mainly on atopic dermatitis since the literature search for acne and psoriasis did not produce results that met our inclusion criteria.
From all the studies taken into consideration, the microorganism mainly responsible for the onset of AD is S. aureus. Moreover, it revealed that a change in some skin physiology parameters occurs in patients with AD. These include an increase in skin pH and TEWL and a decrease in the level of Hydration. But what is the real correlation between these aspects?.
Doubtless, skin's pH is the first factor to be considered responsible for the presence of specific microbial species on the skin. Studies conducted in vitro have shown that higher pH values could favour the colonization of S. aureus [35,36]. Two of the studies taken into consideration have confirmed this aspect, demonstrating that in patients with AD, there is an increase in the skin pH value [26,30].
In patients with AD, there is an increase in TEWL and a decrease in skin hydration. [26-29]. However, neither TEWL nor skin hydration seems to have any direct correlation with the presence of S. aureus. Despite this, the increase in TEWL and the decrease in hydration are associated with an abundance of S. aureus and an increase in its colonization sites [27].
Furthermore, in Baurecht, et al. was analysed the skin’s lipid composition using a mass spectrometer. In this way, was identified an important association between the skin microbiota and the lipid composition. In particular, in one predilection sites of AD, an increase in levels of long-chain unsaturated lipids was correlated with a decrease in bacterial diversity. Moreover, an increase in the levels of ceramides, and in particular of α-hydroxy-ceramides, was connected with an increase in abundance of Staphylococci, and in particular S. aureus.
This could be a first step for the future understanding and the future development of a possible correlation between skin microbiota and physiological barrier parameters.
References
- Drèno B, Araviiskaia E, Berardesca E, Gontijo G, Sanchez Viera M, Xiang L, et al. (2016) Microbiome in healthy skin, update for dermatologists. J Eur Acad Dermatol Venerol 30: 2038-2047.
- Chen YE, Tsao H (2013) The skin microbiome: current perspective and future challenges. J Am Acad Dermatol 69: 143-155.
- Musthaq S, Mazuy A, Jakus J (2018) The microbiome in dermatology. Clinics in Dermatology 36: 390-398.
- Baldwin HE, Bhatia ND, Friedman A,. Eng RM, Seite S (2017) The Role of Cutaneous Microbiota Harmony in Maintaining a Functional Skin Barrier. J Drug Dermatol 16: 12-18.
- Belkaid Y, Segre J (2014) Dialogue between skin microbiota and immunity. Science 346: 954-959.
- Sanford JA, Gallo RL (2013) Functions of the skin microbiota in health and disease. Semin Immunol 25: 370-377.
- Tottè JEE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren E, et al. (2016) Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol 175: 687-695.
- Biedermann T, Skabytska Y, Kaesler S, Volz T (2015) Regulation of T cell immunity in atopic dermatitis by microbes: the Yin and Yang of cutaneous inflammation. Front Immunol 6: 1-9.
- Weyrich LS, Dixit S, Farrer AG, Cooper AJ, Cooper AJ (2015) The skin microbiome: Associations between altered microbial communities and disease. Australas J Dermatol 56: 268-274.
- Avena-Woods C (2017) Overview of atopic dermatitis. Am J Manag Care 23: 115-123.
- McLean WHI, Irvine AD (2012) Heritable Filaggrin Disorders: The Paradigm of Atopic Dermatitis. J Invest Dermatol 132: 20-21.
- Scharschmidt TC, Man M-Q, Hatano Y, Crumrine D, Gunathilake R, et al. (2009) Filaggrin deficiency confers a paracellular barrier abnormality that reduces inflammatory theresholds to irritants and haptens. J Allergy Clin Immunol 124: 496-506.
- Grice E (2014) The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin Cutan Med Surg 33: 98-103.
- Fahlèn A, Engstrand L, Backer B, Powles A, Fry L (2012) Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch Dermatol Res 304: 15-22.
- Owen CM, Chalmers RJG, O'Sullivan T, Griffiths CEM (2001) A systematicreview of antistreptococcal interventions for guttate and chronic plaque psoriasis. Br J Dermatol 145: 886-890.
- Telefer NR, Chalmers RJ, Whale K, Colman G (1992) The role of streptococcal infection in the initiation of guttate psoriasis. Arh Dermatol 128: 39-42.
- Leung DYM, Travers JB, Giorno R, Norris DA, Skinner R, et al. (1995) Evidence for a Streptococcal Superantigen-Driven Process in Acute Guttate Psoriasis. J Clin Invest 96: 2106-2112.
- Balci DD, Duran N, Ozer B, Gunescar R, Onlen Y et al. (2009) High prevalence of Staphylococcus aureus cultivation and superantigen production in patients with psoriasis. Eur J Dermatol 19: 238-242.
- Lynde CW, Poulin Y, Vender R, Bourcier M, Khalil S (2014) Interleukin 17A: Toward a new understanding of psoriasis pathogenesis. J Am Acad Dermato 71: 141-150.
- Fitz-Gibbon S, Tomida S, Chiu B-H, Nguyen L, Du C, et al. (2013) Propionibacterium acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J Invest Dermatol 133: 2152-2160.
- Platsidaki E, Dessinioti C (2018) Recent advances in understanding Propionibacterium acnes (Cutibacterium acnes) in acne. F1000research 7: 1-12.
- Bakry OA, Samaka RM, Sebika H, Seleit I (2014) Toll-like receptor 2 and P. acnes: Do They Trigger Initial Acne Vulgaris Lesions? Anal Quant Cytopathol Histopathol 36: 100-110.
- Beylot C, Auffret N, Poli F, Claudel J-P, Teccia M-T, et al. (2014) Propionibacterium acnes: an update on its role in the pathogenesis of acne. J Eur Acad Dermatol Venereol 28: 271-278.
- Jahns AC, Eliers H, Ganceviviene R, Alexeyev OA (2015) Propionibacterium species and follicular keratinocyte activation in acneic and normal skin. Br J Dermatol 172: 981-987.
- Baurecht H, Rühlemann MC, Rodríguez E, Thielking F, Harder I, et al. (2018) Epidermal lipidic composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J Allergy Clin Immunol 141: 1668-1676.
- Tauber M, Balica S, Hsu C-Y, Jean-Decoster C, Lauze C, et al. (2015) Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol 137: 1272-1274.
- Na S-Y, Roh J-Y, Kim J-M, Tamang MD Lee J-R (2012) Analysis of Colonization and Genotyping of the Exotoxins of Staphylococcus aureus in Patients with Atopic Dermatitis. Ann Dermatol 24: 413-419.
- Bianchi P, Theunis J, Casas C, Villeneuve C, Patrizi A, et al. (2016) Effects of a New Emollient-Based Treatment on Skin Microflora Balance and Barrier Function in Children with Mild Atopic Dermatitis. Pediatr Dermatol 33: 165-171.
- Glatz M, Jo J-H, Kennedy EA, Polley EC, Segre JA, et al. (2018) Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PLOS one 13: 0192443.
- Hülpüsch C, Tremmel K, Hammel G, Bhattacharyya M, de Tomassi A, et al. (2000) Skin pH-dependent Staphylococcus aureus abundance ad predictor for increasing atopic dermatitis severity. Allergy 75: 2888-2898.
- Miajlovic H, Fallon PG, Irvine AD, Foster TJ (2010) Effect of filaggrin breakdown products on growth of and protein expression by Staphylococcus aureus. J Allergy Clin Immunol 126: 1184-1190.
- Hachem J-P, Crumrine D, Fluhr J, Brown BE, Feingold KR (2003) pH Directly Regulates Epidermal Permeability Barrier Homeostasis, and Stratum Corneum Integrity/Cohesion. J Invest Dermatol 121: 245-353.
- Lambers H, Piessens S, Bloem A, Pronk H, Finkel P (2006) Natural skin surface pH is on average below 5, which is beneficial for its resident flora. Int J Cosmet Sci 28: 359-370.
- Barel AO, Clarys P (1995) Study of the stratum corneum barrier function by transepidermal water loss measurements: comparison between twocommercial instruments: Evaporimeter and Tewameter. Skin Pharmacol 8: 186-195.
- Mempel M, Schmidt T, Weidinger S, Schnopp C, Foster T, et al. (1908) Role of Staphylococcus aureus Surface-Associated Proteins in the Attachment to cultured HaCaT keratinocytes in a New Adhesion assay. J Invest Dermatol 111: 452-456.
- Korting HC, Hübner K, Greiner K, Hamm G, Braun-Falco O (1990) Differences in the skin surface pH and bacterial microflora due to the longterm application of synthetic detergent preparations of pH 5.5 and pH 7.0. Results of a crossover trial in healthy volunteers. Acta Dermato Venereol 70: 429-431.
Citation: Tomasoni D, Perugini P (2021) Relationship between Skin Microbiota and Skin Biophysical Parameters in Inflammatory Skin Disease: A Systematic Review. J Clin Dermatol Ther 7: 072.
Copyright: © 2021 Diletta Tomasoni, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

