
The Extracts of Evodiae Fructus and Stephaniae Tetrandrae Radix Show Synergy in Blocking the Entry of SARS-CoV-2
*Corresponding Author(s):
Karl Wah-Keung TsimCenter For Chinese Medicine, The Hong Kong University Of Science And Technology, Clear Water Bay, Kowloon, Hong Kong
Tel:+852 23587332,
Fax:852 23581552
Email:botsim@ust.hk
Abstract
Introduction: COVID-19, caused by the novel coronavirus SARS-CoV-2, has been a constant threat to public health since its outbreak in early 2019. Despite the development of vaccines and therapeutic drugs, the effectiveness of preventing both the infection and recurrence of this respiratory disease is limited due to emerging variants and their side effects. Hence, the development of potent and safe anti-SARS-CoV-2 therapeutic agents is still in urgent demand.
Methods: We previously developed a screening platform comprising various pseudovirus entry tests in conjunction with a computational docking modulation protocol. With this platform, we demonstrated that the extract of Evodia Fructus and its main chemical component, rutaecarpine, inhibited SARS-CoV-2 entry by suppressing the S-protein-ACE2 complex.
Results: Here, we reveal that when the extract of Stephaniae Tetrandrae Radix or its chemical component, tetrandrine (a well-known TPC2 inhibitor), is combined with Evodiae Fructus extract or rutaecarpine, respectively, then these act in a synergistic manner to block the entry of SARS-CoV-2.
Conclusion: Given the fact that herbal extracts have been effectively used in clinical applications and have well-established safety records, our current findings suggest that these drug combinations might be considered as potential anti-COVID-19 treatments.
Keywords
Herbal medicines; Rutaecarpine; SARS-CoV-2; Synergistic anti-COVID-19 effect; Tetrandrine; Variants
Abbreviations
ACE2: Angiotensin-converting enzyme II
DMEM: Dulbecco’s Modified Eagle Medium
EVFEtOH: Evodiae Fructus EtOH extract
HPLC: High-performance liquid chromatography
IC50: Half-maximal inhibitory concentration
PBS: Phosphate-buffered saline
RBD: Receptor binding domain
STREtOH: Stephaniae Tetrandrae Radix EtOH extract
TCM: Traditional Chinese Medicine
TMPRSS2: Transmembrane serine protease 2
TPC2: Two-pore channel subtype 2
Introduction
COVID-19, caused by SARS-CoV-2, has resulted in unprecedent socioeconomic damage around the world, and it is still considered to be a threat to public health. Indeed, as of 22 November 2023, approximately 770 million confirmed cases and 7 million deaths were recorded that were directly related to this disease [1]. Since the start of the pandemic, tremendous efforts have been made to reduce or prevent the risk of infection. For example, a global vaccination program was initiated and over 13 billion doses of various types of vaccine were administrated [1,2]. In addition, several oral drugs were developed, including Molnupiravir from Merck and Paxlovid from Pfizer [3,4]. Nevertheless, SARS-CoV-2 mutated rapidly, and the various mutations, named alpha to omicron from the Greek alphabet, significantly reduced the effectiveness of these treatments. There is therefore still a clear demand for the development of more effective (and safer) drugs to provide alternative clinical options for patients [5,6].
During viral infection, the spike (S)-protein, part of the SARS-CoV-2 virus, binds to a receptor on the host cell called angiotensin-converting enzyme II (ACE2); this leads to virus-host cell fusion and virus invasion. It is known that a specific receptor binding domain (RBD) on the S-protein enables the virus to bind to ACE2 by forming a binding pocket, and so this might prove to be an effective target for inhibitors against viral entry [7-9]. In addition, phosphoinositide is reported to play a crucial role in the entry of SARS-CoV-2 into host cells by regulating endocytosis. PI(3,5)P2 (phosphatidylinositol-3,5-bisphosphate) is one of the phosphoinositides that mediates the maturation of early endosomes to late endosomes, and it can activate two-pore channel subtype 2 (TPC2), which is a member of the voltage-gated ion channel superfamily. Several lines of evidence indicate that when TPC2 function is blocked, then the viral entry of SARS-CoV-2 is inhibited [10,11]. As S-protein and TPC2 are both key factors in driving SARS-CoV-2 entry, we hypothesize that a pharmacological agent that targets both, might block viral entry more efficiently than a drug that targets just one protein residue. This might lead to a reduction in the IC50 and hence the dosage required and side effects, when compared with individual protein inhibitors [12].
During the main outbreak of COVID-19, the Chinese government imposed various measures to fight this deadly disease. One of these was to actively support research on traditional Chinese medicine (TCM). Indeed, several TCM drugs were shown to be effective in relieving the systematic symptoms of COVID-19 as well as curtailing the course of the disease. This helped to control the pandemic. Several TCM prescriptions, including Lianhua Qingwen capsule, Qingfei Paidu Tang, and Huashi Baidu Tang, were recommended by the National Committee of Health in China, as they were shown to have clinical efficacy against SARS-CoV-2 [13]. TCM treatments typically comprise various components that display significant efficacy through different modes of action. The synergistic effect of various TCM components results in better clinical efficacy [14,15]. Thus, it is important to continue to identify other TCMs and their component phytochemicals as potential candidates for the development of new treatments against COVID-19.
In our search for anti-COVID-19 agents, we established a multi-component detecting platform to screen, identify and optimize TCMs and their phytochemical candidates from herbal extracts. The extract of Evodiae Fructus (i.e., the fruit of Evodia rutaecarpa (Juss.) Benth) and its chemical component, rutaecarpine have previously been shown to have an inhibitory effect on SARS-CoV-2 entry into cells by inhibiting the S-protein-ACE2 complex [16]. Moreover, tetrandrine and a tetrandrine-rich Chinese herb, Stephaniae Tetrandrae Radix (Stephania tetrandra S. Moore), have previously been identified as potent TPC2 inhibitors [17,18]. To date, the potential synergistic effect of TPC2 inhibitor plus S-protein inhibitor has not been studied, although the triple combination of anti-viral agents targeting S-protein and transmembrane serine protease 2 (TMPRSS2), has been shown to have a synergistic anti-SARS-CoV-2 effect [19]. This suggests that testing different combinations of various inhibitors might serve as an effective strategy in the design of potent anti-COVID-19 treatments. In light of this, we hypothesize that when S-protein and TPC2 inhibitors are combined, then the viral entry of SARS-CoV-2 is blocked in a synergistic manner.
Material And Methods
- Herbal extract preparation
Evodiae Fructus (i.e., the dried fruit of E. rutaecarpa) and Stephaniae Tetrandrae Radix (i.e., the root of S. tetrandra) were acquired from local herbal retailers and authenticated in accordance with the Hong Kong Chinese Materia Medica Standards [20]. Each herbal powder (10 g) was placed in a round-bottomed flask (250 mL) and dissolved in distilled 90% ethanol (EtOH; 100 mL) to obtain the respective EtOH extract. The solution was then refluxed for 1 h, after which it was filtered through a paper filter with a pore size of 110 µm (Advantec, Tokyo, Japan). The extracts were then dried in a rotary evaporator, and they provided 2.21 g Evodiae Fructus EtOH extract (EVFEtOH) and 1.96 g Stephaniae Tetrandrae Radix EtOH extract (STREtOH) with extraction efficiencies of 22.1% and 19.6%, respectively.
- Cell culture
HEK293T cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with high glucose, which was supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA; hereafter called culture medium) in an incubator at 37°C with a water-saturated atmosphere and 5% CO2. The culture medium was refreshed every other day. HEK293T cells that over expressed human ACE2 (hACE2) were prepared by transfection with a pcDNA3.1-hACE2 plasmid (Addgene, Watertown, MA), and the cell viability was determined, as previously reported [21].
- High-performance liquid chromatography (HPLC) analysis of EVFEtOH and STREtOH extracts
HPLC investigations were conducted according to methods developed by the Hong Kong Chinese Materia Medica Standards [20]. Each herbal extract (1 mg) was placed in a conical flask and dissolved in 50% EtOH (10 mL). Rutaecarpine and tetrandrine (at purities >95% from Chengdu Must, Chengdu, China) were used as HPLC standards and prepared at concentrations of 500 mg/L in 100% EtOH. Each solution was sonicated for 30 min before being filtered through a polytetrafluoroethylene membrane syringe filter (0.45 µm; Anpel Laboratory Technologies, Shanghai, China), after which it was transferred to a 10-mL volumetric flask containing 50% EtOH. In the HPLC analysis, the mobile phase consisted of MilliQ water and acetonitrile (ACN) with the following gradient: 55% ACN (0-20 min); 55–100% ACN (20–30 min); and 100% ACN (30−40 min). The mobile phase flow rate and injection volume were 1.0 mL/min and 10 μL, respectively. The characteristic peaks were detected at wavelengths of 342 nm and 265 nm for rutaecarpine and tetrandrine, respectively.
- SARS-CoV-2 pseudotyped-virus production and host cell entry inhibition
SARS-CoV-2 pseudotyped-virus was produced as described by Lin et al., [21]. ACE2-overexpressing HEK293T cells were seeded into 48-well plates (at a density of 4 x 105 cells/mL), and 400 µL culture medium containing the SARS-CoV-2 pseudovirus (100 µL) ± various treatments were added, after which the mixture was incubated at 37°C for 24 h. This medium was then replenished with fresh culture medium, and the cultures were incubated for a further ~48 h. The cells were then washed with Phosphate-Buffered Saline (PBS), after which a luciferase assay was conducted as previously described [21].
The percentage inhibition of each sample was calculated by the following equation: Inhibition rate (%) = (Luciferase activity of the blank control − Luciferase activity of the detecting sample) / (Luciferase activity of the blank control − Luciferase activity of group in the absence of pseudovirus) × 100%.
A wild-type anti-SARS-CoV-2 neutralizing antibody (A19215, ABClonal, Woburn, MA) and an anti-SARS-CoV-2 omicron antibody (S1N-M122, ACRO, Newark, DE) were used as positive controls (1 µg/mL), whereas a solvent blank (lacking the pseudovirus) was the negative control. The inhibition percentage was calculated based on the luciferase activity without treatments.
- Immunolabeling ACE2
HEK293T cells were cultured on poly-L-lysine-coated coverslips for 48 h, after which they were washed with PBS and fixed with 4% paraformaldehyde for 20 min. The cells were washed three times with PBS and then blocked with PBS containing 0.1% Triton X-100 and 5% bovine serum albumin for 1 h at room temperature. Subsequently, the cells were washed three times with PBS and then they were incubated overnight at 4°C with an anti-ACE2 antibody (code: E-ll, Santa Cruz Biotechnology, Santa Cruz, CA; diluted 1:400) and an anti-Na+/K+ ATPase antibody (code: EP1845Y, Abcam Ltd., Cambridge, UK, diluted 1:400). The cells were then washed with PBS, after which they were incubated with an Alexa Fluor 488-conjugated anti-rabbit antibody and an Alexa Fluor 647-conjugated anti-mouse antibody (Abcam Ltd.) for 2 h at room temperature. The cells were washed with PBS again and then mounted with ProLong® Gold Antifade Reagent containing 4’,6-diamino-2-phenylindole (DAPI; Cell Signaling Technology, Danvers, MA). They were then imaged with a Leica TCS SP8 laser scanning confocal microscope (Leica Microsystems, Germany).
- Immunolabeling TPC2
HEK293T cells were immunolabeled using the method described above for ACE2. However, in these new experiments, cells were incubated with the anti-TPC2 antibody (Covalab, Cambridge, UK; at 1:200 dilution) overnight at 4°C, and then with the Alexa Fluor 488-conjugated anti-rabbit antibody for 2 h at room temperature. The fluorescence labeled cells were mounted using ProLong® Gold Antifade Reagent with DAPI and then imaged with the Leica TCS SP8 confocal microscope.
- Computational calculations using Compusyn
The median-effect equation was conducted with the Compusyn software (https://www.combosyn.com/), accessed on 01/12/2023. This equation is based on the theory that the ratio of the fraction affected (Fa) versus the fraction unaffected (Fu) is equal to the corresponding dosage (D) versus the median-effect dose (Dm) to the mth (sigmodicity of dose-effect curve), where Fa + Fu = 1, and Dm represents the efficacy (Fa/Fu = (D/Dm)m). The combinational index (CI) was also generated from this software, whereby CI < 1, CI = 1, and CI > 1 represent synergistic, additive, and antagonistic effects, respectively.
- Screening of S-protein inhibitors via the Enzyme-Linked Immunosorbent Assay (ELISA)
S-protein inhibition was assessed using a SARS-CoV-2 Spike-ACE2 binding assay kit (ImmunoDiagnostics Ltd. Hong Kong, China) following the manufacturer’s instructions. The reaction was stopped by adding 2 M H2SO4, and data were acquired with a microplate reader (FlexStation; Molecular Devices, San Jose, CA). The percentage of inhibition was calculated using the equation: Percentage of inhibition = (PAvg - SAvg) / PAvg × 100%, where PAvg and SAvg are the mean optical density values of the positive control and test samples, respectively.
- Computational docking analysis
The chemical structures of rutaecarpine and tetrandrine were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/, accessed on 01-12-2023), and the protein structures were downloaded from the Protein Data Bank (https://www.rcsb.org/, accessed on 01-12-2023). Virtual screening was performed using the SEESAR docking software (Version 12.0, https://www.biosolveit.de/, accessed on 01-12-2023) as follows: (i) The binding site of each protein was determined by selecting the corresponding amino acid residues within the binding pocket to form a docking site. Potential ligand binding states, such as protonation and tautomeric forms, were automatically evaluated using the ProToss methodology to generate the most accessible hydrogen network using an internal empirical scoring function: and (ii) A docking modulation of 4.3 × 105 docking clients was subsequently conducted using the “Compute LeadIT Docking” mode of the FlexX algorithm, and ten docking poses for each phytochemical were produced. (iii) The docking energy (ΔG) and estimated HYDE affinity (KiHYDE) for each ligand conformation were calculated using the “Assess Affinity with HYDE in SEESAR” mode in the HYDE rescoring function. The conformation with the lowest HYDE score was determined to be the most favorable for the corresponding ligand [22]. Regarding the docking images, light green indicates that a particular chemical atom is favorable in the docking pocket, whereas light red suggests that the atom is not favorable as it requires relatively high energy to bind.
Results
- HPLC spectra of the herbal extracts
The EtOH extracts of Evodiae Fructus (EVFEtOH) and Stephaniae Tetrandrae Radix (STREtOH) were obtained and shown to have extraction efficiencies of 22.1% and 19.6%, respectively. Subsequent HPLC analysis showed that EVFEtOH and STREtOH contained 0.42% rutaecarpine and 9.71% tetrandrine, respectively (Appendix, Figure S1). These results are very much in line with the criteria described by the Hong Kong Chinese Materia Medica Standards [20].
- EVFEtOH and STREtOH had a synergistic effect on the inhibition of viral entry
Not all cell lines express ACE2 and so these cannot form an S-protein-ACE2 complex. Therefore, we decided to transfect cDNA encoding ACE2 into HEK293T cells to optimize entry of the pseudovirus. As shown in the immunolabeling images in figure 1 (A), after transfection, the cells expressed ACE2 at the cell surface. We also confirmed, via use of an anti-TPC2 antibody, that TPC2 is also expressed in these cells (Figure 1 (B)). 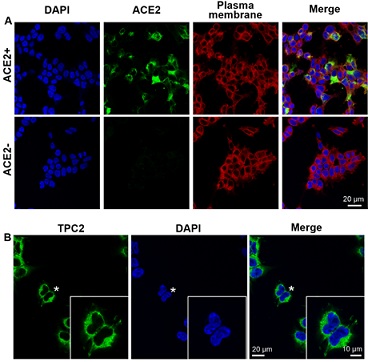
Figure 1: Immunolabeling ACE2 and TPC2 in transfected HEK293T cells. (A) Localization of ACE2 (in green) and the plasma membrane (identified from the localization of Na+/K+ ATPase; in red) in transfected cells. The cell nuclei were stained with DAPI (in blue). The confocal images clearly show that, after transfection, HEK293T cells express ACE2, which permits the formation of the S-protein-ACE2 complex. Representative images are shown, n = 4. ACE2+/-: cells in which the ACE2 receptor was transfected or not. (B) Expression of TPC2 (in green) in HEK293 cells. Again the nuclei are shown in blue. Representative images are shown, n = 4. The cells indicated by the asterisk are shown at a higher magnification in the inset panel.
As demonstrated in our previous manuscript [16], a series of experiments utilizing an S-protein-based ELISA assay was performed to illustrate the anti-viral properties of EVFEtOH and rutaecarpine. A standard inhibitor provided by the supplier (NIBSC code 20/136), was used as a positive control and this showed an S-protein-ACE2 complex inhibition rate of up to 75% (Appendix, Figure S2). Meanwhile, EVFEtOH was found to inhibit the interaction between the S-protein and ACE2 in a dose-dependent manner, with a maximum inhibition of ~100% at ~50 µg/mL (Appendix, Figure S2). Moreover, the major ingredient of EVFEtOH, rutaecarpine, also displayed potent inhibitory activity to the S-protein-ACE2 complex. This suggests that the phytochemical component of EVFEtOH is likely to be responsible (at least to some extent), for the effectiveness of this extract (Appendix, Figure S2). These data suggest that both EVFEtOH and rutaecarpine might act as inhibitors against the S-protein-ACE2 complex and therefore might prevent SARS-CoV-2 entry into host cells.
A pseudovirus entry system, utilizing both wild-type and omicron variants of SARS-CoV-2, was established with the aim of determining the effectiveness of the extracts. The luciferase activity decreased with the dilution of pseudovirus in a virus titering study, indicating that the viral assay was stable for the subsequent studies (Appendix, Figure S3). Next, an MTT assay showed that only slight apoptosis was observed at concentrations over 75 µg/mL for both extracts (Figure 2). STREtOH and its major ingredient tetrandrine are known to act as inhibitors of TPC2 [17,18]. Here, we showed that STREtOH also inhibits the entry of SARS-CoV-2 into host cells (Appendix, Figure S4). As the inhibition of viral entry by EVFEtOH was mediated by the S-protein, and the inhibitory effect of STREtOH was mediated by TPC2, we hypothesized that the two extracts might have a synergy between the two extracts.
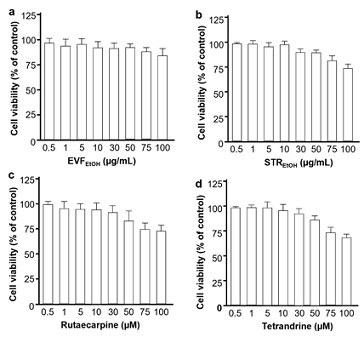
Figure 2: Cell toxicities of EVFEtOH, STREtOH, rutaecarpine and tetrandrine. EVFEtOH and STREtOH were tested at final concentrations of 0.5, 1, 5, 10, 30, 50, 75 and 100 µg/mL, whereas rutaecarpine tetrandrine were tested at final concentrations of 0.5, 1, 5, 10, 30, 50, 75 and 100 µM. MTT solution at 20 μL/well was applied to give a final concentration of 0.5 mg/mL. The optical density of each well was determined at 492 nm. The values are presented as mean ± SD percentage compared to the control (no drug treatments) and n = 4.
We conducted several trials to determine the most efficient ratio of EVFEtOH and STREtOH when used in combination. Intriguingly, when they were used at a 1:1 ratio, then they displayed better inhibitory performance against both the wild-type and omicron pseudovirus than other combination groups (Figure 3). As such, our subsequent synergistic studies were based on a ratio of 1:1 for the two extracts. As expected, EVFEtOH and STREtOH both alone and in combination, showed reasonable potency in the pseudovirus entry assay acting in a dose-dependent manner from 1.5625 µg/mL to 25 µg/mL. In addition, the mixture was found to show a significant synergistic attenuation of viral entry of both wild-type and omicron variants (Figure 4).
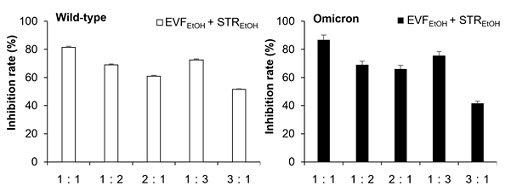 Figure 3: Different combination groups used in the pseudovirus entry tests. EVFEtOH and STREtOH were tested at different ratios in the wildtype (A) and omicron (B) pseudovirus entry test. The ratios of 1:1, 1:2, 2:1, 1:3, 3:1 indicate that the ratio of concentrations (µg/mL) between the two components were, 15:15, 10:20, 20:10, 7.5:22.5, and 22.5:7.5, respectively. The values are presented as mean ± SD percentage of inhibition to the control (no drug treatments) and n = 4.
Figure 3: Different combination groups used in the pseudovirus entry tests. EVFEtOH and STREtOH were tested at different ratios in the wildtype (A) and omicron (B) pseudovirus entry test. The ratios of 1:1, 1:2, 2:1, 1:3, 3:1 indicate that the ratio of concentrations (µg/mL) between the two components were, 15:15, 10:20, 20:10, 7.5:22.5, and 22.5:7.5, respectively. The values are presented as mean ± SD percentage of inhibition to the control (no drug treatments) and n = 4.
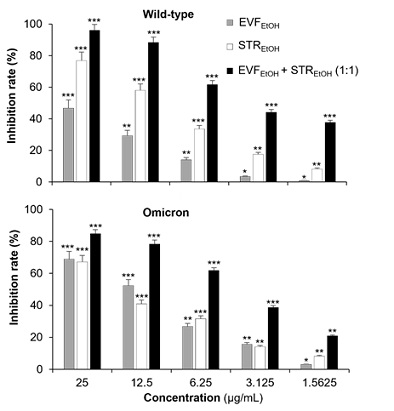
Figure 4: EVFEtOH and STREtOH when used alone or in combination showed dose-dependent anti-viral properties against both the wild-type and omicron SARS-CoV-2 variants. When used alone, EVFEtOH and STREtOH were tested at concentrations of 1.5625, 3.125, 6.25, 12.5 and 25 µg/mL. When used together, EVFEtOH and STREtOH were tested at 1.5625 + 1.5625 µg/mL, 3.125 + 3.125 µg/mL, 6.25 + 6.25 µg/mL, 12.5 + 12.5 µg/mL and 25 + 25 µg/mL. The values are presented as the mean ± SD inhibition rate (%) compared to the control (with no drug treatments) and n = 4.
A computational calculation utilizing the Compusyn software (https://www.combosyn.com/), developed by Dorothy Chou, was employed to establish the median-effect equation and combination index theorem. According to the computational calculation, the median-effect of combining the extracts was higher than the two single extracts alone in the presence of wild-type pseudovirus, with a combination index (CI) less than 1 (Figure 5 (A)). This suggests that when EVFEtOH is applied together with STREtOH, then it has a higher inhibitory activity than when either of the two extracts are used alone. This indicates that when used together, these two extracts have a remarkable synergistic anti-viral effect. A similar scenario was observed in the presence of the omicron variant, as the combined group displayed a higher median-effect than the individual extracts, and the CI value was once again less than 1 when the Fa (fraction unaffected) was less than 0.9 (Figure 5 (B)). This suggests that the synergistic effect was relatively strong even at low concentrations.
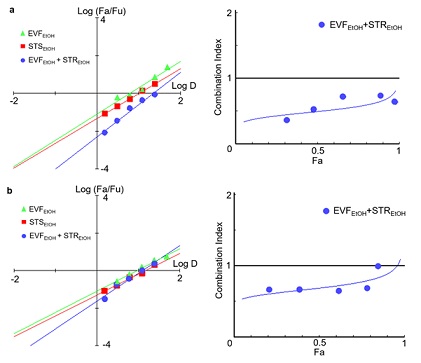
Figure 5: A combination of EVFEtOH and STREtOH had a synergistic inhibitory effect on the (A) wild-type and (B) omicron variants of SARS-Cov-2. The median effect (Log (Fa/Fu)) was calculated according to the fraction affected (Fa) and fraction unaffected (Fu), whereas the combination index was based on the Fa and dose. The median effect and combination index were both generated by the Compusyn software (https://www.combosyn.com/, accessed on 01/10/2023).
- Rutaecarpine and tetrandrine showed a synergistic anti-viral effect
In light of the positive outcome observed when the EVFEtOH and STREtOH extracts were combined, the potential synergistic effect of their corresponding components, (i.e., rutaecarpine and tetrandrine, respectively) was also tested. Similarly, a rutaecarpine and tetrandrine ratio of 1:1 was found to have better efficacy against the wild-type and omicron variants than the other combinations tested (Figure 6). Hence, various concentrations of these chemicals (at a 1:1 ratio) were investigated in the subsequent studies. They were shown to be effective in blocking viral entry of both the wild-type and omicron variants in a dose-dependent manner (Figure 7). It is worth noting, however, that rutaecarpine and tetrandrine when used alone showed very little inhibition at low concentrations (i.e., less than 1 µM), whereas when they were used together, they displayed effective inhibitory activity even at a concentration of 0.625 µM.
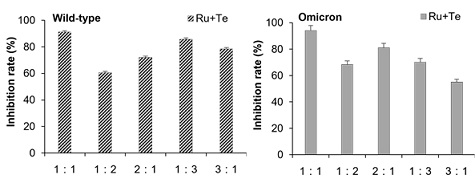
Figure 6: Effect of rutaecarpine (Ru) and tetrandrine (Te) at different combination ratios on the wildtype and omicron pseudovirus entry assay. The ratios of 1:1, 1:2, 2:1, 1:3, and 3:1 indicate that the ratio of concentrations (µM) between the two components were, 15:15, 10:20, 20:10, 7.5:22.5, and 22.5:7.5, respectively. The values are presented as mean ± SD percentage of inhibition to control (no drug treatments) and n = 4.
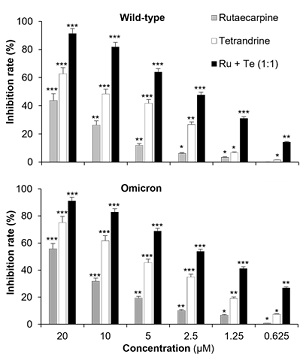 Figure 7: Rutaecarpine (Ru) and tetrandrine (Te) when used alone or in combination showed dose-dependent anti-viral effects against both the wild-type and omicron SARS-CoV-2 variants. When used alone, Ru and Te were tested at concentrations of 1.5625, 3.125, 6.25, 12.5 and 25 µM. When used together, Ru and Te were tested at 1.5625 + 1.5625 µM, 3.125 + 3.125 µM, 6.25 + 6.25 µM, 12.5 + 12.5 µM and 25 + 25 µM. The values are presented as mean ± SD inhibition rate (%) compared to the control (with no drug treatments) and n = 4.
Figure 7: Rutaecarpine (Ru) and tetrandrine (Te) when used alone or in combination showed dose-dependent anti-viral effects against both the wild-type and omicron SARS-CoV-2 variants. When used alone, Ru and Te were tested at concentrations of 1.5625, 3.125, 6.25, 12.5 and 25 µM. When used together, Ru and Te were tested at 1.5625 + 1.5625 µM, 3.125 + 3.125 µM, 6.25 + 6.25 µM, 12.5 + 12.5 µM and 25 + 25 µM. The values are presented as mean ± SD inhibition rate (%) compared to the control (with no drug treatments) and n = 4.
In the subsequent computational calculations, better median-effects were observed in the combined group against both the wild-type (Figure 8 (A)) and omicron variants (Figure 8 (B)), and the CI values were both less than 1. This suggests that rutaecarpine and tetrandrine have a strong synergistic effect in inhibiting viral entry. These findings were very much in agreement with the observations we made in the cell studies (See Section 3.2 and 3.3), which demonstrated that both chemicals were responsible for the effectiveness of their parental herbal extracts.
To further test our hypothesis, we conducted a docking study to detect the potential binding affinities between the phytochemicals and their targeted protein. Indeed, we found that rutaecarpine binds to wild-type (Figure 9 (A)) and omicron (Figure 9 (B)) S-proteins at the RBD, with binding energies of approximately -8.6 kJ/mol and -10.2 kJ/mol, respectively. On the other hand, as expected, tetrandrine anchored at the active site of TPC2 with a predicted binding energy of -9.9 kJ/mol (Figure 9 (C)). In summary, our data indicate that rutaecarpine and tetrandrine can bind to S-protein and TPC2, respectively, which are two key proteins involved in viral entry. Moreover, both phytochemicals showed a significant synergistic effect in blocking viral entry, which suggests that they might contribute to the synergistic anti-viral activities of EVFEtOH and STREtOH.
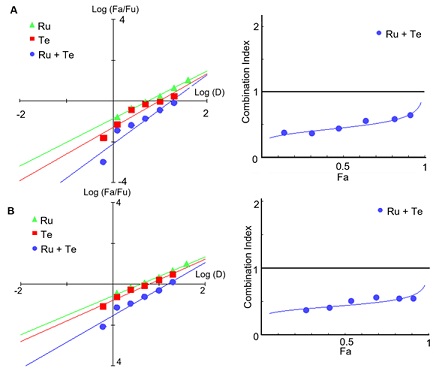
Figure 8: The median-effect of a combination of rutaecarpine (Ru) plus tetrandrine (Te) was higher than when either of these chemical compounds was used alone. Indeed, combination indexes < 1 were calculated, indicating that when used together, rutaecarpine and tetrandrine synergistically suppressed the (A) wild-type and (B) omicron coronavirus. The median effect and combination index were both generated by the Compusyn software (https://www.combosyn.com/, accessed on 01/10/2023).
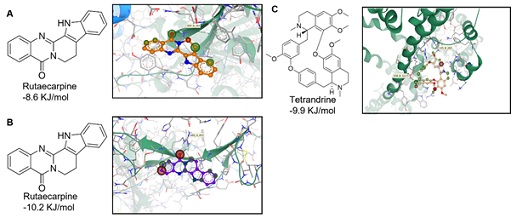
Figure 9: Docking studies of rutaecarpine and tetrandrine against the S-protein and TPC2, respectively. Rutaecarpine was shown to bind to (A) the receptor binding domain (residue 438-506) of the wild-type S-protein (PDB code: 6LZG) with a predicted binding energy of ~ -8.6 KJ/mol and (B) to the omicron S-protein (PDB code: 7T9L) with a binding energy of ~ -10.2 KJ/mol. (C) Tetrandrine was found to bind to the active site of TPC2 (PDB code: 6NQ0) with a binding energy of ~ -9.9 KJ/mol.
Discussion
From a Chinese medicine perspective, COVID-19 is not linked to common pathogenic factors, such as the wind, cold, or dryness, but it is associated with specific epidemic factors, including dampness, heat, toxins, and stasis. As such, a therapeutic agent that can eliminate one or more of these factors might prove to be effective for clinical application [23]. Indeed, several TCM prescriptions have been recommended both by the National Health Commission of China and the World Health Organization to combat the symptoms of COVID-19. For example, Jinhua Qinggan granule and Qingfei Paidu decoction have been utilized to remove heat and clear toxins from the lungs (as described by TCM theory) and both exhibited remarkable effects in relieving the symptoms and shortening the course of recovery. Indeed, in one comprehensive study, these two prescriptions were found to target several key proteins, including 3CL protease, ACE2, BCL2, CASP3, and HSP90AA1. This suggests that the use of multi-target inhibitors against several virus pathogenesis-related proteins that have a close network of interaction might be an efficient strategy for repositioning or repurposing drug candidates against SARS-CoV-2 [24,25].
TCM has been broadly utilized for thousands of years in many countries around the world, and herbal medicines are at the core of TCM, such that more than 10 herbal extracts can be mixed into one single prescription. Intriguingly, the mystery surrounding TCM efficacy was eventually found to be due to the synergistic nature of the various potent herbal extracts or ingredients in a single formulation [26]. Synergy is usually triggered when two or more drugs are combined to generate higher efficacy than the total sum of the individual components. This is unlike an additive effect where the effect of the combined components is equal to the individual agents. Synergistic effects have been demonstrated in several potent treatments for clinical application. For example, Artemisia annua L. and Gardenia jasminoides Ellis (G) exhibit a robust synergistic attenuation of various liver disorders, and oxyresveratrol together with acyclovir exhibit synergistic anti-virus properties against herpes simplex virus [27,28].
Various investigations have demonstrated that when several anti-viral drugs are combined, then such multi-targeted therapeutics are highly effective against SARS-CoV-2 [29-31]. For example, when azithromycin was used with ivermectin, this combination significantly inhibited the viral replication of SARS-CoV-2 [29]. In addition, in a comprehensive high-throughput screening study, when HCV NS5A was combined with remdesivir, then this mixture exhibited remarkable anti-viral synergy against COVID-19 [30]. Furthermore, a combination of pamapimod (an inhibitor of p38 MAPK) and pioglitazone (an anti-inflammatory inhibitor) also triggered synergistic anti-viral activity against SARS-CoV-2 [31]. Our new findings are very much in agreement with these previous reports, and together they serve as strong evidence that combinational treatments can induce synergistic anti-COVID-19 activities.
The dry fruit of E. rutaecarpa, Evodiae Fructus, is well-recorded in the Chinese Pharmacopoeia, and this, or its ingredient rutaecarpine, has been widely utilized (either alone or with other Chinese herbal extracts), to treat gastrointestinal disorders, emesis, dysentery, and postpartum haemorrhage [32]. In addition, Stephaniae Tetrandrae Radix and one of its ingredients, tetrandrine, have been used in clinical applications as diuretics as well as anti-inflammatory and antirheumatic remedies [33]. Here, we revealed that the extract of Evodiae Fructus and rutaecarpine robustly reduced viral infection by inhibiting S-protein to ACE2 binding, and the extract of Stephaniae Tetrandrae Radix and tetrandrine attenuated viral entry via the inhibition of TPC2. Our new data support the findings of previous investigations [16-18]. However, to date, there have been no studies to investigate the synergistic anti-viral effect of Evodiae Fructus and Stephaniae Tetrandrae Radix together, and so our new data are the first to demonstrate the synergistic effect of this combination. Our in vitro investigations now pave the way for follow-up pharmacodynamic and pharmacokinetic studies.
We also demonstrated the potential of S-protein and TPC2 inhibitors as synergistic anti-viral agents. This provides a novel strategy for the design of other promising anti-SARS-CoV-2 agents. Given the fact the Evodiae Fructus and Stephaniae Tetrandrae Radix along with their respective ingredients (rutaecarpine and tetrandrine), have all been employed as clinical treatments for several years, we strongly believe that our new findings indicate that these are promising candidates for the development of new potent therapeutics against COVID-19.
Conclusion
COVID-19 has resulted in millions of deaths worldwide and it continues to cause socioeconomic damage in many countries. Here, we demonstrated that the S-protein inhibitors, EVFEtOH and rutarcarpine, and TPC2 inhibitors, STREtOH and tetrandrine, expressed robust anti-viral activity by preventing the entry of the SARS-CoV-2 pseudovirus into host cells. Interestingly, we also revealed that if EVFEtOH was combined with STREtOH, or if rutaecarpine was combined with tetrandrine, then these had a significant synergistic effect in blocking viral entry. Such combination groups might therefore be promising anti-COVID-19 candidates.
Author’s Contribution
Conceptualization, KW-KT and ALM; methodology, SL and XW; software, SL; validation, KWL; formal analysis, RD.; investigation, RW-LT and TK.; resources, TT-XD; writing—original draft preparation, SL; writing—review and editing, SEW, ALM and KW-KT; project administration, KWL and SEW. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a Health and Medical Research Fund (COVID-19) award (HMRF20SC07(COVID190213)) from the Hong Kong Government Food and Health Bureau; Hong Kong RGC Theme-based Research Scheme (T13-605/18-W); TUYF19SC02, PD18SC01 and HMRF18SC06; The Key-Area Research and Development Program of Guangdong Province (2020B1111110006); Hong Kong Innovation Technology Fund (PRP/076/20FX; Hong Kong RGC-GFC 16100921; Shenzhen Science and Technology Innovation Committee (ZDSYS201707281432317); Zhongshan Municipal Bureau of Science and Technology (2019AG035); GBA Institute of Collaborate Innovation (GICI-022); Special Project of Foshan University of Science and Technology in 2019 (FSUST19-SRI10); PRP/076/20FX; UIM/385, ITS/500/18FP; ITCPD/17-9); HMRF20SC07; AFD20SC01; and Guangzhou Science and Technology Committee Research Grant (GZSTI16SC02; GZSTI17SC02).
Institutional Review Board Statement: Not applicable
Informed Consent Statement: Not applicable.
Data Availability Statement: All original data can be obtained from the corresponding author upon request.
Acknowledgment: We thank Mr. Hung Chun Lee and Miss Wan Suet Yip for their contributions to this work.
Conflicts of Interest: The authors declare no conflict of interest.
Appendix Supplementary Figures
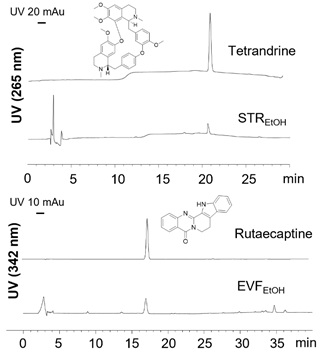
Figure S1: HPLC analysis. The HPLC spectra of tetrandrine (0.5 mg/mL) in STREtOH (0.1 mg/mL) with UV absorption of 265 nm (upper panels), and rutaecarpine (0.5 mg/mL) in EVFEtOH (0.1 mg/mL) with UV absorption of 342 nm (lower panels). Injection volumes of 10 µL were used.
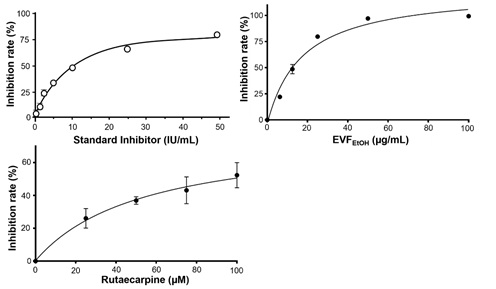
Figure S2: Effect of EVFEtOH and rutaecarpine in an S-protein-based ELISA assay. A standard inhibitor, provided by the supplier (calibrated to NIBSC code 20/136), showed good potency in this assay. In addition, EVFEtOH and rutaecarpine both robustly suppressed interactions between the S-protein and ACE2 in a dose-dependent manner. The values are presented as the mean ± SD percentage of inhibition to the control (with no drug treatment) and n = 4.
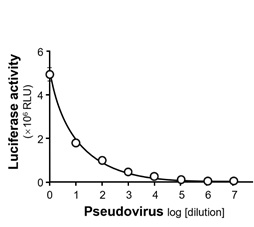
Figure S3: The intracellular luciferase activity (indicating the rate of pseudovirus entry into host cells) was determined at different dilutions of wild-type pseudovirus. RLU: relative light units.
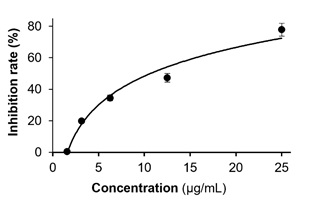
Figure S4: Wild-type pseudovirus entry test of STREtOH extract. STREtOH was tested at concentrations of 1.5625, 3.125, 6.25, 12.5 and 25 µg/mL. The values are presented as mean ± SD percentage of inhibition to control (no drug treatments), and n = 4.
References
- World Health Organization (2024) Number of COVID-19 cases reported to WHO. World Health Organization, Geneva, Switzerland.
- McMenamin ME, Nealon J, Lin Y, Wong JY, Cheung JK, et al. (2022) Vaccine effectiveness of two and three doses of BNT162b2 1 and CoronaVac against COVID-19 in Hong Kong. The Lancet Infectious Diseases 22:1435-1443.
- Sanderson T, Hisner R, Donovan-Banfield I, Hartman H, Løchen A, et al. (2023) A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes. Nature 623: 594-600.
- Liu J, Pan X, Zhang S, Li M, Ma K, et al. (2023) Efficacy and safety of Paxlovid in severe adult patients with SARS-Cov-2 infection: a multicenter randomized controlled study. The Lancet Regional Health - Western Pacific 33:100694.
- Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, et al. (2022) SARS-CoV-2 omicron variant: antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science 375:760-764.
- Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, et al. (2021) SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409-424.
- Shang J, Wan Y, Luo C, Ye G, Geng Q, et al. (2020) Cell entry mechanisms of SARS-CoV-2. PNAS 117:11727-11734.
- Gil C, Ginex T, Maestro I, Nozal V, Barrado-Gil L, et al. (2020) COVID-19: drug targets and potential treatments. J Med Chem 63:12359-12386.
- Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, et al. (2020) SARS-CoV-2: Structure, biology, and structure-based thera-peutics development. Front Cell Infect Microbiol 10: 587269.
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. (2020) Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11: 1620.
- Moccia F, Negri S, Faris P, Perna A, Luca AD, et al. (2021) Targeting endolysosomal two-pore channels to treat cardiovascular disorders in the novel coronavirus disease 2019. Front Physiol 12: 629119.
- Makhoba XH, Viegas C Jr, Mosa RA, Viegas FPD, Pooe OJ (2020) Potential impact of the multi-target drug approach in the treatment of some complex diseases. Drug Des Devel Ther 14: 3235-3249.
- Kang X, Jin D, Jiang L, Zhang Y, Zhang Y, et al. (2022) Efficacy and mechanisms of traditional Chinese medicine for COVID-19: a systematic review. Chin Med 17: 30.
- Li Z, Chen H, Zhang H, Li Y, Wang C, et al. (2020) Similarity and specificity of traditional Chinese medicine formulas for management of coronavirus disease 2019 and rheumatoid arthritis. ACS omega 5: 30519-30530.
- Choudhry N, Zhao X, Xu D, Zanin M, Chen W, et al. (2020) Chinese Therapeutic strategy for fighting COVID-19 and potential small-molecule inhibitors against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J Med Chem 63: 13205-13227.
- Lin S, Wang X, Guo H, Dai N, Tang RW, et al. (2023) The ethanol extract of Evodiae Fructus and its ingredient, rutaecarpine, inhibit infection of SARS-CoV-2 and inflammatory responses. Int J Mol Sci 24: 762.
- Li K, Chen X, Zhang J, Wang C, Xu Q, et al. (2022) Transcriptome Analysis of Stephania tetrandra and Characterization of Norcoclaurine-6-O-Methyltransferase Involved in Benzylisoquinoline Alkaloid Biosynthesis. Front Plant Sci 13: 874583.
- Sakurai Y, Kolokoltsov AA, Chen CC, Tidwell MW, Bauta WE, et al. (2015) Two-pore channels control Ebolavirus host cell entry and are drug targets for disease treatment. Science 347: 995-998.
- Wagoner J, Herring S, Hsiang TY, Ianevski A, Biering SB, et al. (2022) Combinations of host- and virus-targeting antiviral drugs confer synergistic suppression of SARS-CoV-2. Microbiol Spectrum 10: 0333122.
- HKCMS Office (2011) Hong Kong Chinese Medicine Medica Standards. Department of Health: The Government of Hong Kong, Special Administrative Region, Hong Kong.
- Lin S, Wang X, Tang RW, Lee HC, Chan HH, et al. (2022) The extracts of Polygonum Cuspidatum Root and Rhizome block the entry of SARS-CoV-2 wild-type and omicron pseudotyped viruses via inhibition of the S-protein and 3CL protease. Molecules 27: 3806-3819.
- Spagnolli G, Massignan T, Astolfi A, Biggi S, Rigoli M, et al. (2021) Pharmacological inactivation of the prion protein by targeting a folding intermediate. Commun Biol 4: 62-77.
- Luo H, Gao Y, Zou J, Zhang S, Chen H, et al. (2020) Reflections on treatment of COVID-19 with traditional Chinese medicine. Chin Med 15: 94.
- Guan W, Lan W, Zhang J, Zhao S, Ou J, et al. (2020) COVID-19: Antiviral agents, antibody development and traditional Chinese medicine. Virologica Sinica 35: 685-698.
- Kreutzberger AJB, Sanyal A, Ojha R, Pyle JD, Vapalahti O, et al. (2021) Synergistic block of SARS-CoV-2 infection by combined drug inhibition of the host entry factors PIKfyve kinase and TMPRSS2 protease. Virol J 95: 0097521.
- Joshi RS, Jagdale SS, Bansode SB, Shankar SS, Tellis MB, et al. (2021) Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn 39: 3099-3114.
- Zhou X, Seto SW, Chang D, Kiat H, Razmovski-Naumovski V, et al. (2016) Synergistic Effects of Chinese Herbal Medicine: A Comprehensive Review of Methodology and Current Research. Front Pharmacol 7: 201.
- Ren PX, Shang WJ, Yin WC, Ge H, Wang L, et al. (2022) A multi-targeting drug design strategy for identifying potent anti-SARS-CoV-2 inhibitors. Acta Pharmacologica Sinica 43: 483-493.
- Forni DD, Poddesu B, Cugia G, Chafouleas J, Lisziewicz J, et al. (2022) Synergistic drug combinations designed to fully suppress SARS-CoV-2 in the lung of COVID-19 patients. PLoS ONE 17: 0276751.
- Nguyenla X, Wehri E, Dis EV, Biering SB, Yamashiro LH, et al. (2022) Discovery of SARS-CoV-2 antiviral synergy between remdesivir and approved drugs in human lung cells. Sci Rep 12: 18506.
- Setz C, Große M, Auth J, Fröba M, Rauch P, et al. (2022) Synergistic antiviral activity of pamapimod and pioglitazone against SARS-CoV-2 and its variants of concern. Int J Mol Sci 23: 6830.
- Xiao SJ, Xu XK, Chen W, Xin JY, Yuan WL, et al. (2023) Traditional Chinese medicine Evodiae Fructus: botany, traditional use, phytochemistry, pharmacology, toxicity and quality control. Nat Prod Bioprospecting 13: 6.
- Jiang Y, Liu M, Liu H, Liu S (2020) A critical review: traditional uses, phytochemistry, pharmacology and toxicology of Stephania tetrandra Moore (Fen Fang Ji). Phytochem Rev 19: 449-489.
Citation: Lin S, Wang X, Tang RW-L, Leung KW, Duan R, et al. (2024) The Extracts of Evodiae Fructus and Stephaniae Tetrandrae Radix Show Synergy in Blocking the Entry of SARS-CoV-2. J Altern Complement Integr Med 10: 478.
Copyright: © 2024 Shengying Lin, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

