
Detection of Extended Spectrum b-lactamases Producing Genes among Third Generation Cephalosporins Sensitive Bacterial Strains from a Medical College Hospital in Bangladesh
*Corresponding Author(s):
Taslima YesminDepartment Of Microbiology, TMSS Medical College, Bogra, Bangladesh
Tel:01713 377003,
Email:taslimasanta@yahoo.com
Abstract
Methods: This cross sectional study was undertaken in non-repetitive clinical isolates collected from Mymnesingh Medical College Hospital, Mymensingh, Bangladesh from both the outpatient and inpatient departments over a period of six months from January 2011 to June 2011. The ESBL status was confirmed by double disc diffusion test and minimum inhibitory concentration by agar dilution method as recommended by Clinical and Laboratory Standard Institute 2010 and multiplex polymerase chain reaction for TEM, SHV and CTX-M genes.
Results: A total of 300 Gram negative bacilli were included in the study; among them 236 were resistant and 64 were sensitive to 3GCs by disc diffusion test. Multidrug resistant ESBL production was found 75.8% from resistant isolates and 54.6% from sensitive isolates. Rate of TEM, SHV and CTX-M genes present among sensitive strains were 36.4%, 24.2% and 21.1%, respectively.
Conclusion: Both common and new ESBLs genes were detected from 3GCs sensitive bacteria of which TEM is most common. Routine ESBLs screening for all Gram-negative isolates both from sensitive and resistant to 3GCs strains might be useful for the physicians in selecting effective antibiotics.
Keywords
INTRODUCTION
Phenotypic methods are not able to distinguish between the specific enzymes responsible for ESBLs production. The determination of whether a specific ESBL present in a clinical isolate is related to TEM and SHV enzymes is a complicated process because point mutations around the active sites of the TEM and SHV sequences have led to amino acid changes that increase the spectrum of activity of the parent enzymes, such as in TEM-1, TEM-2, and SHV-1 [3]. The molecular method commonly used is the PCR amplification of the genes with oligonucleotide primers, followed by sequencing. Sequencing is essential to discriminate between the non-ESBL parent enzymes to different variants of TEM or SHV of ESBLs [8].
Prevalence of ESBL producing from Escherichia coli and Klebsiella pneumoniae isolates were 41% and 36% in Pakistan [9] and 43.2% and 39.5%, respectively in Bangladesh [10]. Another study conducted in a medical college hospital, Bangladesh found 41.66% ESBL in Gram-negative bacteria [11]. A number of studies in India have reported as ESBL producers were 40.85%, 51.4% and 53.84% in 2004, 2007 and in 2010, respectively [12-14]. However, another two studies in Iran and India documented high prevalence 96% and 97%, respectively [15,16]. All of these studies were conducted among 3GCs resistant strains. A wide variation of those reports suggests that detection of ESBLs also need to find out among 3GCs sensitive strains. The ESBL genes detected in 3GCs bacterial strains is challenging for physician to treat. Since no previous data is available on the prevalence of genes responsible for ESBLs production in Bangladesh, it is urgent to know whether the bacteria posses the ESBL gene or not and what type of gene they carry. Therefore, the study aimed to detect ESBL genes among the third generation cephalosporin sensitive bacterial strains in Bangladesh.
MATERIALS AND METHODS
A total of 300 non-repetitive clinical isolates were collected from MMCH both the outpatient and inpatient departments over a period of 6 months from January 2011 to June 2011. Urine and skin wound (pus, wound infection) were used as specimen. Laboratory work was carried out in the department of microbiology in MMC. Specimens were collected aseptically. All samples were routinely cultured on MacConkey and blood agar plates at 37°C aerobically for 18 hours. Gram negative isolates were further characterized by standard biochemical tests [17]. The susceptibility to antibiotics was determined by Kirby Bauer method on Muller Hinton agar (MHA) according to Clinical and Laboratory Standard Institute (CLSI) protocols for Gram negative panels [18].
Isolates were screened for ESBL production by using disc diffusion test on MHA media according to the CLSI recommendations where isolates showing inhibition zone size of ≥22 mm with ceftazidime (30 μg), ≥25 mm with ceftriaxone (30 μg), ≥27 mm with cefotaxime (30 μg) , ≥27 mm with aztreonam (30 µg) were suspected for ESBL production [19] (Figure 1).
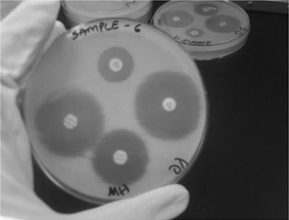
Figure 1: Double disc diffusion test.
Plasmid DNA was isolated from bacterial cells by alkaline lyses method by Medici et al., [20]. A single colony of each organism was inoculated from Mac Conkey agar into 5 ml of Luria-Bertanii broth (LB) and incubated for 20 h at 37°C. Cells from 1.5 ml of the overnight culture was harvested by centrifugation at 12,000 rpm for 5 min. 1.5 ml from LB media containing cells was taken eppendorf tube, than 100 µl TNE buffer was mixed. The mixture was centrifuged for 1 min at 10000 rpm and supernatant was discarded. Again 100 µl NaOH (50 mM) was added to pellet. After heating at 40°C in water bath for 1 min, 60 µl of IM Tris HCl (pH 6.7) was added. Vortex, centrifuge at 10000 rpm for 1 min was done. Then supernatant was used as template (1 µl) (Medici et al. 2003 with some modification). Supernatant 1 µl was used as template. The PCR primers and cycling conditions used were previously described (Table 1) [21-23].
| Target gene | Primers | Sequences | Amplicon size |
| TEM | TEM F | CTTCCTGTTTTTGCTCACCCA | 717 bp |
| TEM R | TACGATACGGGAGGGCTTAC | ||
| SHV | SHV F | TCAGCGAAAAACACCTTG | 471 bp |
| SHV R | TCCCGCAGATAAATCACC | ||
| CTX-M-U | CTX-MU1 | (5? ATGTGCAGYACCAGTAARGT) | 593 bp |
| CTX-MU2 | (5? TGGGTRAARTARGTSACCAGA) |
Table 1: Primer for TEM, SHV, CTX-M-U were used.
Preparation of reaction mixture for PCR amplification, 1 µl of template DNA was added to 50 µl of master mixture containing 4 µl of dNTP mixture (2.5 mM of each), 10X PCR buffer 5 µl (Ex Taq), 0.5 µl of Taqpolymerase (250 U), 1 µl of each primer stock solution (50 pmol/µl), and remaining 38.5 µl volume was fulfilled by nuclease free water (Takara Japan).
Denaturation at 94°C for 30 sec
Annealing at 50°C for 30 sec
Extension at 72°C for 2 min
Final extension at 72°C for 10 minutes
For CTX-M gene
Denaturation at 94°C for 50 sec
Annealing at 50°C for 40 sec
Extension at 72°C for 1 min
Final extension at 72°C for 5 minutes

Amplification was performed in an Eppendorf Thermocycler (Takara, Japan) in 1.0% agarose gel and visualized by staining with ethidium bromide (0.5 mg/ml), running through horizontal electrophoresis. A 100 bp ladder molecular weight marker (Roche, USA) was used to measure the molecular weights of amplified products. The images of ethidium bromide stained DNA bands were digitized using a gel documentation system (AlphaimagerTM 3400, USA) by UV light (Figure 2).
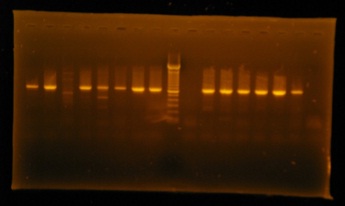
RESULTS
During the 6 months study period a total of 300 various bacterial isolates were yielded. The specimens were urine 216 (72%), wound swab 45 (15%), pus 39 (13%). Out of 300 Gram negative isolates in this study more than half (52%) were Escherichia coli followed by Proteus spp. (18.3%), Klebsiella spp. (15%), Pseudomonas spp. (3%) and others for example Enterobacter spp., Citrobacter spp. were 11.7%. Among the Gram negative isolates ESBLs producers were 179/236 (75.8%) from resistant isolates and 35/64 (54.6%) from sensitive (Table 2). Rate of TEM, SHV and CTX-M genes present in 3GCs sensitive strains were 36.4%, 24.2% and 21.1%, respectively. Among them single strain contain three genes and five strains contain two combine genes (Table 3).
| 3 GCs | ESBL Positive n (%) |
ESBL Negative n (%) |
Total n (%) |
| Sensitive | 35 (54.6) | 29 (45.3) | 64 (100.0) |
| Resistance | 179 (75.8) | 57 (24.1) | 236 (100.0) |
| Total | 214 (71.3) | 86 (28.7) | 300 (100.0) |
Table 2: Distribution of ESBL producing Bacteria according to Third Generation Cephalosporin Sensitivity (n=300).
*χ2 value 11.02 at df 3; p<0.001: Highly Significant; 3GCs: Third Generation Cephalosporins.
| Name of Gene | Frequency | Percentage |
| TEM gene | 12 | 36.4 |
| SHV gene | 8 | 24.2 |
| CTX-M gene | 7 | 21.2 |
| Combined gene | 6 | 18.2 |
| Total | 33 | 100.0 |
DISCUSSION
Our study documented that among the 3GCs sensitive strains more than half (54.6%) were phenotypic positive for ESBLs production (Table 2). Most of the strains were sensitive to ceftazidime, ceftriaxone, and cefotaxime disc but some were only to cefotaxime. They showed more resistant to ceftazidime, which is supported by a study of SHV-3 like ESBLs found 100.0% resistant to ceftriaxone where as only 50.0% were resistant to ceftazidime in vitro [30]. Moland et al., found the sensitivity of using ceftazidime resistance as marker for ESBL production to be 78.0% in a broth dilution format [31]. Similarly we found statistically more significant by these combination. Though ceftazidime plus clavulinic acid (CAZ/CAZC) was the best single disc diffusion test was recommended by George et al., [32] but it cannot cover all the strain. Use of only one combination may fail to detect ESBL positive strains and thus might cause low prevalence [10]. The use of cefotaxime, ceftazidime, and ceftriaxone as the only indicator of ESBLs screening can no longer be recommended. If only one indicator antibiotic would be used for screening, cefpodoxime has proven to be the best molecule for screening all types ESBLs producers in clinical sample, that also produce AmpC β-lactamase [33]. In this study we used two combinations with clavulanic acid (CAZ/CAZC and CTX/CTXC) and found that Escherichia coli and Klebsiella spp. showed maximum ESBLs production in CAZ/CAZC combination, which correlates with other studies [32,34]. But the combination of ceftazidime and cefotaxime still missed two strains producing the SHV-5 and SHV-7 ESBLs [31].
Occurrence and distribution of ESBLs differs from country to country and from hospital to hospital [35]. Since no previous data is available about the prevalence of genes responsible for ESBLs production in Bangladesh, it is assumed that this high rate of ESBLs by phenotypic method may be due to mutation of first two parent gene TEM-1, SHV-1 and newer most prevalent gene CTX-M in the world and our neighbor country India [28]. CTX-M gene is now the most common in Escherichia coli in community and it may be due to overuse of ceftriaxone or due to fecal carriage and transfer gene by horizontal transmission [3,6,27,36]. The present study detected TEM, SHV and CTX-M genes in 36.4%, 24.2%, and 21.1% from phenotypically confirmed ESBLs producers, respectively. Moreover, presence of multiple genes is also found in these bacteria.
Our study should be viewed in the context of several limitations. Firstly, because of constraints of time and budget, this was a single centered study with relatively a small sample; therefore the results cannot be generalized to the whole country. Finally, we did not take into consideration of antibiotic history of the patients before and after in vitro sensitivity test, and we cheeked only in vitro drug response. However, the study represents the first study to determine the presence of ESBL genes from the 3GCs sensitive gram-negative bacterial strains in Bangladesh. Further study need to be conducted to correlate between in vitro and in vivo drug response among such a patients, and to compare antibiotic usage with less or no antibiotic usage.
In conclusion, it can be said that the 3GCs sensitive bacteria possessed both common and newer ESBL genes of which TEM is most common. By considering the fact that Bangladesh is a common place of irrational use and abuse of antibiotics, routine ESBLs screening for all Gram-negative isolates both from sensitive and resistant to 3GCs sensitive bacterial strains might be helpful for the physicians in selecting effective antibiotic therapy.
ACKNOWLEDGMENTS
REFERENCES
- Andrews J (2009) Detection of extended spectrum ß lactamases (ESBLs) in Escherichia coli and Klebsiella species. British society for antimicrobial chemotherapy 674-675.
- Chaudhary U, Aggarwal R (2004) Extended spectrum -lactamases (ESBL)-an emerging threat to clinical therapeutics. Indian J Med Microbiol 22: 75-80.
- Perez F, Endimiani A, Hujer KM, Bonomo RA (2007) The continuing challenge of ESBLs. Curr Opin Pharmacol 7: 459-469.
- Matsumoto Y, Ikeda F, Kamimura T, Yokota Y, Mine Y (1988) Novel plasmid-mediated beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob Agents Chemother 32: 1243-1246.
- Gruteke P, Goessens W, Van Gils J, Peerbooms P, Lemmens-Den Toom N, et al. (2003) Patterns of resistance associated with integrons, the extended-spectrum beta-lactamase SHV-5 gene, and a multidrug efflux pump of Klebsiella pneumoniae causing a nosocomial outbreak. J Clin Microbiol 41: 1161-1166.
- Pitout JD, Laupland KB (2008) Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8: 159-166.
- Sasirekha B, Manasa R, Ramya P, Sneha R (2010) Frequency and Antimicrobial Sensitivity Pattern of Extended Spectrum ß- Lactamases Producing Escherichia coli and Klebsiella pneumoniae Isolated In A Tertiary Care Hospital. Al Ameen J Med Sci 3: 265-271.
- Livermore DM, Hawkey PM (2008) CTX-M: changing the face of ESBLs in the UK. 2005, defining an extended-spectrum beta-lactamase. Clin Microbiol Infect 5: 21-24.
- Jabeen K, Zafar A, Hasan R (2005) Frequency and sensitivity pattern of Extended Spectrum beta Lactamase producing isolates in a tertiary care hospital laboratory of Pakistan. J Pak Med Assoc 55: 436-439.
- Rahman MM, Haq JA, Hossain MA, Sultana R, Islam F, et al. (2004) Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in an urban hospital in Dhaka, Bangladesh. Int J Antimicrob Agents 24: 508-510.
- Haque R, Salam MA, Abdus MD, Salam (2010) Detection of ESBL producing nosocomial gram negative bacteria from a tertiary care hospital in Bangladesh. Pak J Med Sci 26: 887-891.
- Babypadmini S, Appalaraju B (2004) Extended spectrum -lactamases in urinary isolates of Escherichia coli and Klebsiella pneumoniae-prevalence and susceptibility pattern in a tertiary care hospital. Indian J Med Microbiol 22: 172-174.
- Sharma S, Bhat GK, Shenoy S (2007) Virulence factors and drug resistance in Escherichia coliisolated from extraintestinal infections. Indian J Med Microbiol 25: 369-373.
- Yusha’u M, Kumurya AS, Aliyu HM, Suleiman L (2010) Prevalence of Extended spectrum ß lactamases among Enterobacteriaceae in Murtala Mohammed specialist hospital, Kano, Nigeria. Bayero Journal of Pure and Applied Sciences 3: 169- 172.
- Naehi L, Shahcheraghi F, Nikbin VS, Nematzadeh S (2010) PER, CTX-M, TEM and SHV Beta lactamases in Clinical Isolates of Klebsiella pneumoniae Isolated from Tehran, Iran. Iran J Basic Med Sci 13: 111-118.
- Sharma J, Sharma M, Ray P (2010) Detection of TEM & SHV genes in Escherichia coli & Klebsiellapneumoniae isolates in a tertiary care hospital from India. Indian J Med Res 132: 332-336.
- Chessbrough M (2006) District laboratory practice in tropical countries. Part-2, 2nd edn, Cambridge University, Newyork, USA pp: 184- 186.
- Clinical Laboratory Standards Institute (2010) Performance standards for antimicrobial disc susceptibility testing. 14th informational supplement, USA.
- CLSI (2010) Performance Standards for Antimicrobial Susceptibility Testing, Twentieth Informational Supplement, CLSI Document M100-S20, Wayne, PA, USA.
- De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, et al. (2003) Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella entericaserotype enteritidis in poultry. Appl Environ Microbiol 69: 3456-3461.
- Pagani L, Dell'Amico E, Migliavacca R, D'Andrea MM, Giacobone E, et al. (2003) Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J Clin Microbiol 41: 4264-4269.
- Chia JH, Chu C, Su LH, Chiu CH, Kuo AJ, et al. (2005) Development of a multiplex PCR and SHV melting-curve mutation detection system for detection of some SHV and CTX-M beta-lactamases of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae in Taiwan. J Clin Microbiol 43: 4486-4491.
- Lal P, Kapil A, Das BK, Sood S (2007) Occurrence of TEM & SHV gene in Extended Spectrum Beta-Lactamases (ESBLs) producing Klebsiella sp. isolated from a tertiary care hospital. Indian J Med Res 125: 173-178.
- Tenover FC, Mohammed MJ, Gorton TS, Dembek ZF (1999) Detection and reporting of organisms producing extended-spectrum beta-lactamases: survey of laboratories in Connecticut. J Clin Microbiol 37: 4065-4070.
- Steward CD, Wallace D, Hubert SK, Lawton R, Fridkin SK, et al. (2000) Ability of laboratories to detect emerging antimicrobial resistance in nosocomial pathogens: a survey of project ICARE laboratories. Diagn Microbiol Infect Dis 38: 59-67.
- Karas JA, Pillay DG, Muckart D, Sturm AW (1996) Treatment failure due to extended spectrum beta-lactamase. J Antimicrob Chemother 37: 203-204.
- Ensor VM, Shahid M, Evans JT, Hawkey PM (2006) Occurrence, prevalence and genetic environment of CTX-M beta-lactamases in Enterobacteriaceae from Indian hospitals. J Antimicrob Chemother 58: 1260-1263.
- Peirano G, Pitout JD (2010) Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35: 316-321.
- Cavaco LM, Abatih E, Aarestrup FM, Guardabassi L (2008) Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur, or cefquinome. Antimicrob Agents Chemother 52: 3612-3616.
- Farkosh MS (2007) Extended-Spectrum beta lactamase Producing Gram Negative Bacilli.
- Moland ES, Black JA, Ourada J, Reisbig MD, Hanson ND, et al. (2002) Occurrence of newer beta-lactamases in Klebsiella pneumoniae isolates from 24 U.S. hospitals. Antimicrob Agents Chemother 46: 3837-3842.
- Jacoby GA, Munoz-Price LS (2005) Mechanisms of disease: The New beta-Lactamases. N Engl J Med 35: 380-391.
- Black JA, Thomson KS, Buynak JD, Pitout JD (2005) Evaluation of beta-lactamase inhibitors in disk tests for detection of plasmid-mediated AmpC beta-lactamases in well-characterized clinical strains of Klebsiella spp. J Clin Microbiol 43: 4168-4171.
- Thomson KS, Sanders CC, Washington JA 2nd (1991) High-level resistance to cefotaxime and ceftazidime in Klebsiella pneumoniae isolates from Cleveland, Ohio. Antimicrob Agents Chemother 35: 1001-1003.
- Ali AM, Abbasi BSA, Ahmed M (2009) Frequency of Extended Spectrum Beta Lactamase (ESBL) Producing Nosocomial Isolates In A Tertiary Care Hospital In Rawalpindi. Pak Armed Forces Med J.
- Cavaco LM, Abatih E, Aarestrup FM, Guardabassi L (2008) Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur, or cefquinome. Antimicrob Agents Chemother 52: 3612-3616.
Citation: Yesmin T, Hossain A, Paul SK, Yusuf A, Sultana S, et al., (2014) Detection of Extended Spectrum β-lactamases Producing Genes among Third Generation Cephalosporins Sensitive Bacterial Strains from a Medical College Hospital in Bangladesh. J Allergy Disord Ther 1: 001.
Copyright: © 2014 Taslima Yesmin, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

