
A modified CTAB protocol for Genomic DNA extraction from Citrus spp. (Orange)
*Corresponding Author(s):
Muhammad Imran QadirDepartment Of Molecular Biology And Biotechnology, Bahauddin Zakariya University, Multan, Pakistan
Tel:+92 0308 6752329,
Email:mi742813@gmail.com
Abstract
The revolution in the best growth and high yield of plants and crops is due to genetic modification. We explain a simple and efficient method for the genomic DNA extraction from citrus woody plants. Harvesting of fresh leaves from orange plants is done as it contains low level of poly phenyl. DNA extraction is relatively easy from citrus woody than non-citrus plants. CTAB is used in this method of DNA extraction. Ethanol and NaCl are used to remove plants polysaccharides. Same volume of isopropanol is added into supernatant to make DNA pellet. Use 70% ethanol to wash DNA pellet. Then pellet dissolve in TE buffer. 50-500micro g/g DNA pellet is used as a sample. Then PCR & RFLP analysis are carried out. This DNA then can be stored for long durations. Changes and modifications can also be done to get the best quality and high yield from woody plants as well as all kinds of crops.
Keywords
CTAB; DNA pellet; Genomic DNA extraction; PCR; RFLP, TE buffer
INTRODUCTION
In Watson & F Crick had made a structural model of DNA. They suggested that DNA consists of nitrogen bases (A, T, G, C), Phosphate backbone and De-oxy ribose sugar. Historical background of pants and crops shows that cross breeding was mostly adapted method to make changes in their variety. Genomic DNA extraction is the first step for studying the molecular and genetic aspects of any organism. DNA extraction from plant sources is difficult due to the large amount of polysaccharides and poly-phenols which become problematic during PCR reactions. Different protocols were used for genomic DNA extraction but CTAB is most commonly used with modifications [1]. After the discovery of DNA, a broad field was introduced to extract DNA from plants as well as different crops. This is due to find out the genetic differences among the same species of plants. After the extraction of DNA from orange plants, it has become easy to find out the differences of activated genes in one variety but not in other one. The development and application of sampling and DNA extraction protocols include adaptations to plain procedures to suit various inoculum sources. Soil is taken into account to be a posh environment and dealing with DNA recovered from soil is commonly problematic due to the presence of PCR-inhibiting chemical components in soil. Therefore, PCR-based studies require extensive DNA extraction methods and careful purification of macro molecule extracts so as to get rid of humic acids and other contaminants [2]. This possibility, therefore, compels the difference of techniques to sensitize detection. The employment of a two-step PCR with nested primers increases both the sensitivity and also the specificity of the PCR significantly, which might overcome problems with relatively impure DNA extracts and low pathogen numbers.
It is essential to extract high quality DNA during work on it. Extraction of DNA from woody plants is not easy as they contain high polysaccharides level. But in case of citrus plants, DNA extraction is relatively easy. A simple but important protocol is used to extract DNA from Citrus spp. A proper sequence of steps is present in DNA extraction through CTAB method. CTAB, extraction buffer, liquid nitrogen, chloroform, ethanol, isopropanol, 70% ethyl alcohol, NaCl, TE Buffer, PCR & RFLP are the steps and protocol used in this DNA extraction method [3]. For getting best quality DNA, all steps which are arranged in a proper sequence must be followed.
METHODS AND MATERIALS
Plant material
Fresh leaves were harvested from citrus plants (orange) from bio-park in BZU Multan. Then these leaves were kept in ice box to make them safe from degradation. Fresh leaves were selected as they contain low level of polysaccharide (Figure1).

Figure1: Fresh leaves were selected as they contain low level of polysaccharide.
Equipments and Reagents
- • Mortar & pestle
- • Centrifuge
- • Micro-pipettes
- • Eppendorf tubes
- • Water bath
- • Liquid Nitrogen
- • Extraction buffer 100 mM Tris-HCl (pH 8)
- • mM NaCl
- • 50 mM EDTA(pH 8)
- • 5% B-mercaptoethanol
- • 5% (W/V) CTAB
- • Chloroform-isoamyl alcohol (24:1)
- TE buffer(pH 8)
- • 10mM Tris-HCl
- • 1 mM EDTA
- • 10 mg/mL RNase
- • 70% ethanol
- • 5 M NaCl
- • Agarose
DNA Extraction Protocol
- • Grind 5-8 g of fresh leaves of orange tree with pestle &mortar by using liquid nitrogen(or EDTA)
- • Transfer fine powder into 50 ml centrifuge tube and add 15ml extraction buffer
- • Gently invert and put all the tubes in water bath
- • Put off from water bath. Add 15 ml chloroform isoamyl- alcohol (24:1) and invert the tubes gently
- • Centrifuge at 9000 rpm for 10 min
- • Transfer upper layer in another clean tube while debris is discard out
- • Add equal volume of isopropanol , gently invert then put the tubes in incubator at -20 for 30 min
- • Again centrifuge at 9000 rpm for 10 min
- • Supernatant is discarded while lower precipitates remain left
- • Wash the DNA pellets with 3 ml 70% ethanol (2- 3 times)
- • Air dry the pellets for 20-30 min
- • Add 3ml TE buffer & 10 mg/ml RNase
- • Incubate at 37C
- • Transfer DNA solution in fresh tube and add 3 ml phenol- chloroform isoamyl alcohol(25:24:1)
- • Gently invert and again centrifuge at 9000 for 15 min
- • Supernatant transfer into clean tube, add 1 ml of 5M NaCl and 4 ml ether in it
- • Centrifuge at 9000 rpm for 15 min
- • Supernatant is discarded, bottom layer is carefully transferred into clean tube
- • Add chilled isopropanol and remove the DNA pellet with a hook
- • Wash the DNA pellets via 70%ethanol
- • Add 1ml TE buffer to dissolve the DNA pellet
- • Dilute 15 micro L DNA solution for qualitative analysis
- • Add 5 micro L DNA solution in each sample to test on 0.8% agarose
- • Store the DNA solution at -20C
Gel Documentation System & DNA Display
A complete procedure is followed to make agarose gel which act as carrier for DNA. Gel documentation is a system interlinked to computer used to display the proper sequencing of DNA.
RESULTS AND DISCUSSION
There are a lot of methods which are use to extract DNA but this protocol of DNA extraction is cost effective and efficient. Removal of polysaccharides from citrus plants leaves is easy by using this method. As the DNA pellets formed then quickly dissolve in TE buffer so this thing is a good way to the protection of DNA from degradation [4]. Low level of polysaccharides in citrus fruits plants (e.g. orange) is helpful for isolating high quality DNA via PCR and PFLP (Figure 2). When DNA obtained from Citrus plants leaves treated with 0.8% agarose gel, the sample present in the well appeared bright under UV light [5] 70% ethanol play an important for the proper washing of DNA pellets to remove the debris. Citrus spp. mostly preferred for DNA extraction as these species are treated by using CTAB DNA extraction method (Figure 3).
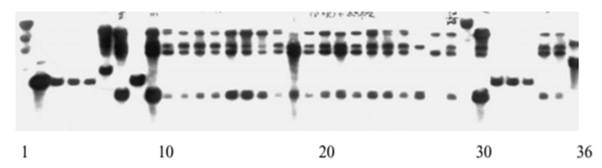 Figure 2: Low level of polysaccharides in citrus fruits plants (e.g. orange) is helpful for isolating high quality DNA via PCR and PFLP.
Figure 2: Low level of polysaccharides in citrus fruits plants (e.g. orange) is helpful for isolating high quality DNA via PCR and PFLP.
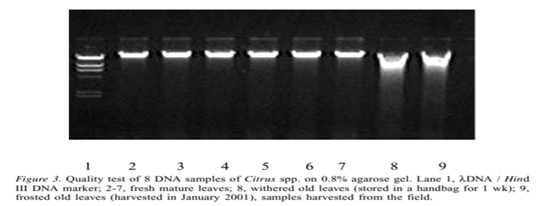 Figure 3: Quality test of 8 DNA samples of Citrus spp. On 0.8% agarose gel. Lane 1, DNA/ Hind III DNA marker; 2-7, fresh mature leaves; 8, withered old leaves (stored in a handbag for 1 week); 9, frosted old leaves (harvested in January 2001), samples harvested from the field.
Figure 3: Quality test of 8 DNA samples of Citrus spp. On 0.8% agarose gel. Lane 1, DNA/ Hind III DNA marker; 2-7, fresh mature leaves; 8, withered old leaves (stored in a handbag for 1 week); 9, frosted old leaves (harvested in January 2001), samples harvested from the field.
CONCLUSION
There are many hurdles to extract DNA from plants but in most cases accumulated amount of polysaccharides is responsible to resist against qualitative DNA extraction [6-10]. SDS (Sodium Dodecyl Sulfate (SDS) is an important chemical which is used for cleaning purpose. In case of DNA extraction, SDS is used for cell lysis. It is also used for denaturing of proteins in electrophoresis in SDS-Page (figure 4).
Figure 4: To overcome this problem, Citrus fruit plants are selected for the best qualitative DNA extraction. Due to low level of polysaccharides, Citrus fruit plants are mostly use for DNA extraction.
AUTHOR’S CONTRIBUTIONS
Muhammad Ishfaq, Faraz and Jahangeer planned and designed the research; Nasir, Sania Javeed and Subhan performed the experiments; Muhammad Ishfaq, Fatima Zahra, Furqan and Momina Batool analysed the data; Muhammad Ishfaq wrote the manuscript.
ACKNOWLEDGEMENT
This research was financially supported by Bahauddin Zakariya University, Multan, Pakistan. I am thankful to Prof. Dr. Muhammad Baber and Dr. Muhammad Imran Qadir for reading and criticizing the draft of this manuscript.
REFERENCES
- Qadir MI, Malik SA (2010) Comparison of alterations in red blood cell count and alterations in hemoglobin concentration in patients suffering from rectal carcinoma undergoing 5-fluorouracil and folic acid therapy. Pharmacologyonline 3: 240-243.
- Qadir MI, Noor A (2018) Anemias. Rare & Uncommon Diseases. Cambridge Scholars Publishing. Newcastle, England.
- Qadir MI, Javid A (2018) Awareness about Crohn’s Disease in biotechnology students. GloAdv Res J Med Medical Sci 7: 62-64.
- Qadir MI, Ishfaq S (2018) Awareness about hypertension in biology students. Int J Mod Pharma Res, 7(2): 08-10.
- Qadir MI, Shahzad R (2018) Awareness about obesity in postgraduate students of biotechnology. Int J Mod Pharma Res 7: 14-16.
- Qadir MI, Saba G (2018) Awareness about intestinal cancer in university student. Nov Appro in Can Study 1: 15-16.
- Qadir MI, Saleem A (2018) Awareness about ischemic heart disease in university biotechnology students. GloAdv Res J Med Medical Sci 7: 59-61.
- Qadir MI, Mehwish (2018) Awareness about psoriasis disease. Int J Mod Pharma Res, 7(2): 17-18.
- Qadir MI, Rizvi M (2018) Awareness about thalassemia in post graduate students. MOJ Lymphology&Phlebology 2: 14-16.
- Qadir MI, Ghalia BA (2018) Awareness survey about colorectal cancer in students of M. Phil Biotechnology at BahauddinZakariya University, Multan, Pakistan. Nov Appro in Can Study 1: 10-11.
Citation: Qadir MI, Ishfaq M (2020) Leucocytes proper Concentration in Urine is necessary for good Human Health. Biotech Res Biochem 3: 010
Copyright: © 2020 Muhammad Ishfaq, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

