
Bio Prospecting Plant Growth-Promoting Bacteria Isolated from Maize (zea mays l.) Roots
*Corresponding Author(s):
Lygia Vitória Galli TerasawaDepartment Of Genetics, Federal University Of Parana, Curitiba, Brazil
Tel:+55 4133611772,
Email:lgterasawa@gmail.com
Abstract
Maize (Zea mays L.) culture has a great importance in several countries, especially in Brazil the third-largest world producer. The increase in maize production has been achieved with a high use of fungicide; however, in view of a more sustainable agriculture plant growth promoting bacteria have been explored aiming for the replacement of chemical fertilizers and biological control. In this study, we investigated the bacterial community isolated from maize roots in order to evaluate their capacity of growth promotion as well as of inhibition of fungal species associated with maize leaf diseases. All isolates evaluated were positive for at least one of the parameters evaluated-growth promotion, enzymatic production or bio control. The best results were observed for Enterobacter sp. LGMB221 and Bacillus sp. LGMB242 that showed the high potential for growth promotion, acting in the early stage of maize seedlings development. Bacillus sp. LGMB152 showed the best enzymatic results, indicating that it might play a role against pathogens, a premise supported by the antagonist activity observed. The next steps involve evaluations under field conditions to confirm if these isolates have biotechnological potential as inoculants for the maize crop. In addition, we suggest that Enterobacter strains LGMB221 and LGMB235 and Escherichia strain LGMB159 might represent new species, indicating the high diversity of bacteria in maize rhizosphere that remains to be determined.
Keywords
16S rRNA; Fungi antagonism; PGPB
INTRODUCTION
Especially in the past few years, the focus of many research groups is to find Plant Growth-Promoting Bacteria (PGPB) that will act in plant growth through one or more mechanisms, including biological nitrogen fixation, phosphate solubilization, production of hormones such as auxins, modifying root diameter [2,3].
Besides the use as PGPB, bacteria have attract attention for their potential to inhibit phytopathogens development, bean alternative to the application of fungicides, mitigating the environmental impacts and contributing to a more sustainable agriculture [4]. Studies performed with tomatoes (Solanum lycopersicum) [5], soybean (Glycine max) and cotton (Gossypium arboretum) [6] have shown that PGPB can act as antagonists to others pathogenic strains, been an effective biological controller, and in the same time increasing grain production. However, the selection of an effective PGPB consists of annex tensive and indispensable preliminary biochemical analysis in vitro. Despite the high amount of studies conducted in plant growth promoting, we decided to explore a previous culture collection of bacteria isolated from different maize genotypes including maize lineages and their respective hybrids [7], in order to evaluated if these isolates can act as plant growth promoting in a different hybrid from the one that they were isolated.
In this way, the aim of this study was to identify bacterial strains isolated from roots of different maize genotypes and to characterize their potential to be used as plant growth promoter and fungal biological controller by in vitro and in vivo evaluations.
MATERIAL AND METHODS
Strains and 16S rRNA gene sequencing
Plant growth promotion evaluation
Analysis of siderophore production was carried out according to Schwyn and Neil [16] adapted by using solid DYGS medium (2g dextrose; 1.5g peptone; 2g yeast extract; 0.5g K2HPO4; 0.5g MgSO4; 1.5g L-glutamic acid; 15g agar 15g, pH 7.0) adding CAS (60.5g of chromo azurol S into 50mL of distilled water plus 10mL of FeCl3.6H2O 1mM in HCl 10mM) carefully mixed into 72.9mg of HDTMA (Hexadecyltrimethylamonium) dissolved in 40mL of distilled water. Phosphate solubilization was evaluated according to Chagas Junior et al., [17], using the culture medium GL (10g glucose; 2g yeast extract; 15g agar added to 0.25g/L of K2HPO4 solution and 1g/LCaCl2, pH 6.5). Positive result was revealed by the halo formation around the colony.
Biological nitrogen fixation was evaluated as described by Araújo et al., [18], using the JNFb semi-solid medium (5g malic acid; 0.6g K2HPO4; 1.8g KH2PO4; 0.2g MgSO4.7H2O; 0.1g NaCl; 0.02g CaCl2.2H2O; 20mg yeast extract; 0.08mg CuSO4.5H2O; 2.4mg ZnSO4.7H2O; 2.8mg H3BO3; 2mg Na2MoO4.2H2O; 2.35mg MnSO4.H2O; 65.6mg Na2 EDTA; 0.1mg biotin; 0.2mg pyridoxine; 4.5g KOH; 2mL bromothymol blue 0.5%; 2.2g agar, pH 6.8) [19]. The bacteria growth was revealed by the formation of a pellicle on the medium surface.
Indol Acetic Acid (IAA) production was evaluated using the methodology described by Kuss et al. [20], modified by using DYGS culture medium containing 10µL of tryptophan 10mg/mL. Salkowski solution (FeCl3.6H2O 2% + H2SO4 37%) was added to reveal the results, absorbance values were measured by spectrophotometry at 530nm wavelength and final values were expressed in µg/mL. Correlation data for IAA production and seed germination was performed using Bio Estat 5.0 [21].
Seed germination was evaluated using the commercial hybrid maize SX2530 provided by Semilia Genética e Melhoramento Ltda, in order to evaluate the interaction of the isolated bacteria with a different hybrid from the one that they were isolated. Seeds were superficially disinfested by immersion in 70% ethanol (v/v) for one minute, three minutes in sodium hypochlorite 3% (v/v), 30 seconds in 70% ethanol (v/v) and then washed three times in sterilized distilled water for one minute. The experiment was performed using 48 seeds for each isolate. The bacteria were growth in LB culture medium. Microbiolization was done by adding the seeds to the culture medium containing 108cells/mL during two hours at 37°C. Microbiolized seeds were placed in germination paper humidified with distilled water and incubated in Biochemical Oxygen Demand (BOD) at 28°C for seven days [22]. Evaluations of length (cm) and volume (cm3) of roots and hypocotyls were verified using Win-Rhizov.4.0 software (Regent Systems, Quebec, Canada). For statistical analysis, Kruskal-Wallis test (p < 0.05) was performed by Assistat 7.6 Beta [23].
Enzymatic profile
Urease production was evaluated in urease culture medium (0.5g Na2HPO4; 0.5g K2HPO4, 0.5g; 0.2g MgSO4.7H2O; 10mL NaCl 10%; 1g yeast extract; 2.5mL bromothymol blue 0.5%, pH 5,8) [29]. Positive result was revealed by the change of culture medium to blue color.
Fungi antagonism
STATISTICAL ANALYSIS
The analysis of correlation between IAA production and the seed germination were performed using the Pearson correlation test, considering hypocotyl and root growing, at Bio Estat 5.0 software [21].
RESULTS
Bacteria identification
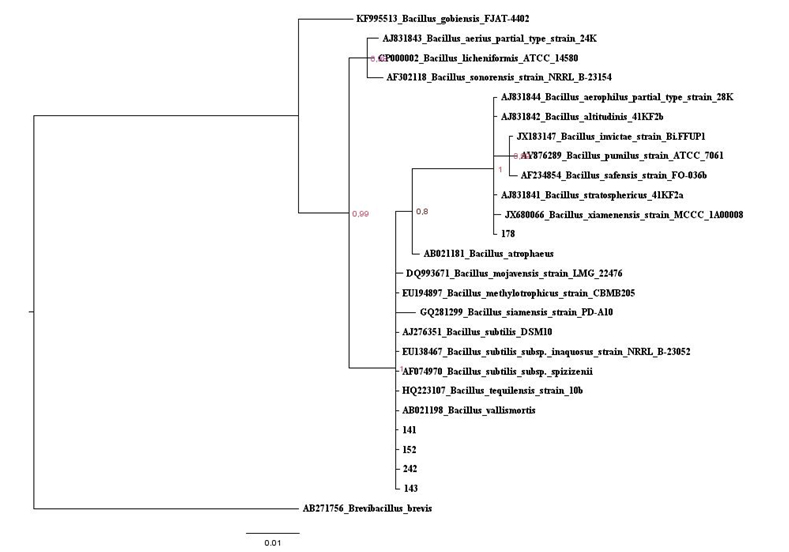 Figure 1: Baysian inferencetree based on the 16S rRNA gene of maize isolates and Bacillus type strains belonging to Clade 1. The species Brevibacillusbrevis was used as outgroup. Values on the node indicate bootstrap support. Bar indicates 10 substitutions per 1,000 nucleotides.
Figure 1: Baysian inferencetree based on the 16S rRNA gene of maize isolates and Bacillus type strains belonging to Clade 1. The species Brevibacillusbrevis was used as outgroup. Values on the node indicate bootstrap support. Bar indicates 10 substitutions per 1,000 nucleotides. Figure S1: Maximum likelihood tree based on the 16S rRNA gene of maize isolates and Bacillus type strains, in red is the isolates that belonging to Clade 1. The species Brevibacillusbrevis was used as out group. Values on the node indicate bootstrap support. Bar indicates 2 substitutions per 1,000 nucleotides.
Figure S1: Maximum likelihood tree based on the 16S rRNA gene of maize isolates and Bacillus type strains, in red is the isolates that belonging to Clade 1. The species Brevibacillusbrevis was used as out group. Values on the node indicate bootstrap support. Bar indicates 2 substitutions per 1,000 nucleotides.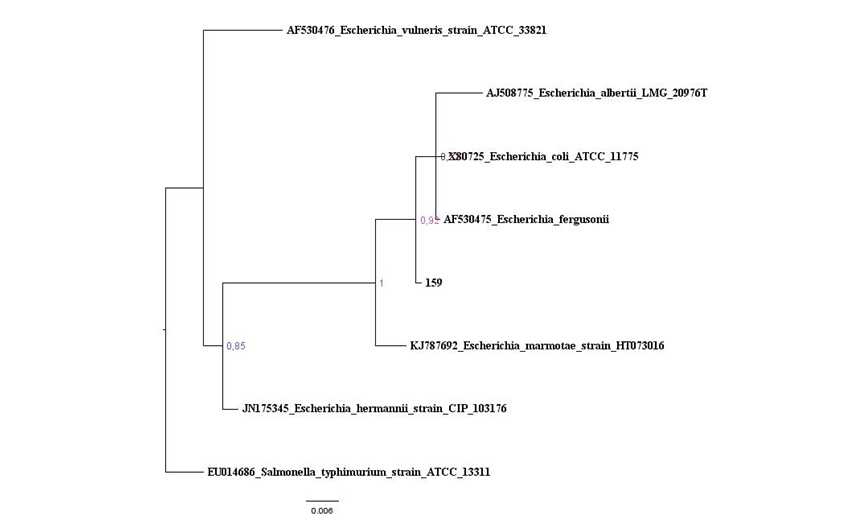 Figure 2: Baysian Inference tree based on 16S rRNA gene of maize isolates and Escherichia species. Salmonella thyphimurium was used as outgroup. Values on the node indicate bootstrap support. Bar indicates 6 substitutions per 1,000 nucleotides.
Figure 2: Baysian Inference tree based on 16S rRNA gene of maize isolates and Escherichia species. Salmonella thyphimurium was used as outgroup. Values on the node indicate bootstrap support. Bar indicates 6 substitutions per 1,000 nucleotides.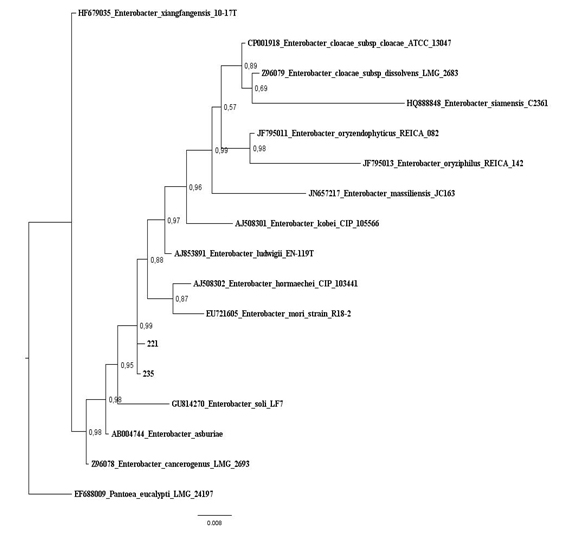 Figure 3: Baysian Inference tree based on 16S rRNA gene of maize isolates and Enterobacter species. Pantoea eucalypti was used as outgroup. Values on the node indicate bootstrap support. Bar indicates 8 substitutions per 1,000 nucleotides.
Figure 3: Baysian Inference tree based on 16S rRNA gene of maize isolates and Enterobacter species. Pantoea eucalypti was used as outgroup. Values on the node indicate bootstrap support. Bar indicates 8 substitutions per 1,000 nucleotides.Characterization for plant growth promotion
|
Strain Code |
GenBank Accession Number |
Identification Based on 16S rRNA Phylogeny |
IAA(µg/mL) |
BNF |
Siderophore |
Phosphate Solubilization |
Amylase |
Cellulase |
Lipase |
Pectinase |
Protease |
Chitinase |
Urease |
|
LGMB 141 |
KY848228 |
Bacillus sp. |
2.16 |
- |
+ |
- |
- |
- |
- |
- |
+ |
- |
- |
|
LGMB 143 |
KY848229 |
Bacillus sp. |
25.75 |
- |
+ |
- |
- |
- |
- |
+ |
- |
- |
- |
|
LGMB 152 |
KY848230 |
Bacillus sp. |
2.54 |
- |
+ |
- |
- |
+ |
+ |
+ |
+ |
- |
+ |
|
LGMB 159 |
KY848231 |
Escherichia sp. |
13.43 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
LGMB 178 |
KY848232 |
Bacillus sp. |
7.29 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
LGMB 221 |
KY848233 |
Enterobactersp. |
7.29 |
- |
+ |
- |
- |
- |
- |
- |
- |
+ |
- |
|
LGMB 235 |
KY848234 |
Enterobactersp. |
14.24 |
- |
+ |
- |
- |
- |
+ |
- |
- |
+ |
+ |
|
LGMB 242 |
KY848235 |
Bacillus sp. |
7.59 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
We also evaluated the capacity of the isolates in affect ingmaize seed development (Table 2). Bacillus sp. LGMB242 increased root volume by 25.7% and root length by 34.9%, in comparison with the control (without bacteria inoculation); the other strains did not show any improvement in the root length or volume. Enterobacter sp. LGMB221 increased hypocotyl length and volume by 21.8% and 33.2%, respectively, but no effect was observed by the other strains.
|
Inoculated Bacteria |
Root Length (mm) |
Root Volume (mm3) |
Hypocotyl Length (mm) |
Hypocotyl Volume (mm3) |
|
Control without bacteria |
372.13CD |
1.94BCD |
718.95CD |
23.51CD |
|
Bacillus sp. LGMB143 |
42.16A |
0.48A |
563.36BCD |
15.18ABCD |
|
Bacillus sp. LGMB152 |
370.14BCD |
2.11CD |
299.68ABC |
8.35ABC |
|
Escherichia sp. LGMB159 |
145.73ABC |
1.26ABC |
364.63ABCD |
12.88ABCD |
|
Bacillus sp. LGMB178 |
136.01AB |
0.91AB |
237.30A |
2.13A |
|
Enterobacter sp. LGMB221 |
261.80ABCD |
1.69ABCD |
492.49ABCD |
15.99BCD |
|
Enterobacter sp. LGMB235 |
225.64ABCD |
1.35ABCD |
875.89D |
31.32D |
|
Bacillus sp. LGMB242 |
296.50ABCD |
1.86ABCD |
262.30AB |
7.29AB |
|
Bacillus sp. LGMB141 |
467.92D |
2.61D |
350.45ABCD |
11.43ABCD |
About extracellular enzymes production, Enterobacter sp. LGMB235 produced lipase and urease, Bacillussp. LGMB143 produced pectinase and LGMB141 protease. Bacillus sp. LGMB152 produced the four enzymes and cellulase either. Chitinase was produced by isolates LGMB221 and LGMB235 (both classified as Enterobacter sp.).
Antifungal activity
|
Straincode |
ID16S RNA |
LGMF1021 |
LGMF1044 |
LGMF1046 |
LGMF1047 |
LGMF1048 |
LGMF1054 |
|
Alternariasp. |
Colletotrichumgraminicola |
Fusarium verticillioides |
Cercosporazeae-maydis |
Bipolarismaydis |
Diaporthesp. |
||
|
LGMB141 |
Bacillus sp. |
1.57A |
3.40C |
2.67B |
2.47B |
3.63B |
3.53B |
|
LGMB143 |
Bacillus sp. |
0.57A |
1.67ABC |
1.27AB |
0.60AB |
3.50B |
3.10AB |
|
LGMB152 |
Bacillus sp. |
0.63A |
1.10AB |
1.07A |
0.83AB |
2.23AB |
1.33A |
|
LGMB159 |
Escherichia sp. |
0.57A |
1.77ABC |
1.10A |
0.53A |
0.93A |
1.33A |
|
LGMB178 |
Bacillus sp. |
0.57A |
1.50ABC |
1.80AB |
1.17AB |
2.57AB |
2.57AB |
|
LGMB221 |
Enterobactersp. |
1.27A |
1.60ABC |
2.00AB |
1.27AB |
2.37AB |
2.07AB |
|
LGMB235 |
Enterobactersp. |
0.60A |
0.87A |
2.23AB |
0.57A |
1.80AB |
2.40AB |
|
LGMB242 |
Bacillus sp. |
0.53A |
2.03BC |
2.17AB |
1.23AB |
2.57AB |
3.33AB |
|
Isolated Strain |
Phytopathogeninhibition in % |
|||||
|
Alternaria sp. |
Colletotrichum |
Fusarium |
Cercosporazeae-maydis |
Bipolaris maydis |
Diaporthe sp. |
|
|
Bacillus sp. LGMB141 |
63.91 |
50.98 |
52.56 |
75.71 |
3.58 |
12.18 |
|
Bacillus sp. LGMB143 |
59.66 |
67.65 |
60.05 |
66.26 |
38.48 |
62.23 |
|
Bacillus sp. LGMB152 |
63.91 |
48.04 |
58.8 |
78.41 |
74.29 |
62.23 |
|
Escherichia sp. LGMB159 |
63.91 |
55.88 |
32.58 |
52.77 |
29.29 |
27.29 |
|
Bacillus sp. LGMB178 |
19.32 |
52.94 |
25.09 |
48.72 |
34.8 |
41.45 |
|
Enterobacter sp. LGMB221 |
61.78 |
74.51 |
16.35 |
77.06 |
50.41 |
32.01 |
|
Enterobacter sp. LGMB235 |
66.03 |
40.2 |
18.85 |
50.07 |
29.29 |
5.57 |
|
Bacillus sp. LGMB242 |
66.03 |
53.92 |
23.85 |
64.91 |
48.58 |
33.9 |
DISCUSSION
Among the 150 bacterial isolates obtained in our previous study, eight were selected for PBPB ability as well as antagonism properties based on the variability observed in a BOX PCR analysis [7]. Strain LGMB159 was identified as Escherichia sp., but probably representing a new species and the same was observed for Enterobacter spp. strains (LGMB221 and LMGB235). Isolates belonging to Enterobacteriaceae family (Enterobacter, Escherichia, Pantoea) have been commonly described as plant-associated bacteria, and previously studies showed a high ability of these generato produce indoles, such as IAA that has an important aspect in plant growth promotion [32-34]. In addition, bacteria belonging to genus Bacillus are common associated with plant growth promotion due the production of different factors, such as, IAA, siderophores, HCN and ammonia [35-37]. These elements are crucial for agricultural crops, wild plants and microalgae [38-41].
Siderophores production act stimulating plant growth by multiple mechanisms, including the provision of iron to plants, production of phytohormones and organic acids that can act solubilizing phosphate. These mechanisms can act direct and indirectly as making nutrients available for plant absorption or depriving pathogenic organisms of essential elements to survival [34]. In our study, strains LGMB141, LGMB143, LGMB152, LGMB221, and LGMB235 showed siderophores production, that besides acting in iron assimilation by plants, can also act as an iron kidnapper from phytopathogens [37]. However, none of the isolates evaluated showed the ability to solubilize phosphate or fixing nitrogen.
All strains produced considerable amounts of IAA, with an emphasis to Bacillus sp. LGMB143; followed by Escherichia sp. LGMB159 and Enterobacter sp. LGMB235. The amount of IAA produced by these strains is substantially higher than in other studies with bacteria of the same genus [42,43]. However, the results are alike to that reported under similar environmental conditions from this region [44], which may suggest that these strains are able to produce IAA by the same biosynthetic pathways. Considering the effects on seed germination, Bacillus sp. LGMB242 increased root length and volume, while Enterobactersp. LGMB221 increased hypocotyl length and volume. Harsh et al., [45], found that some microbial strains can produce bio stimulants such as auxins from precursors existing in plants roots, or IAA production. However, in our study, strains that increased the root and hypocotyl development are not the strains with the highest IAA production in vitro. This data can be explained by once the addition of microbial auxin can change the optimal level of endogenous auxin and can cause plant growth inhibition [46].
Production of extracellular enzymes by microorganisms plays an important role in plant pathogens control, in addition to other biotechnological applications [47]. About the enzymes evaluated in our study, just two strains (Bacillus sp. LGMB178 and Escherichia sp. LGM159) showed no extracellular enzyme production, but one strain (Bacillus sp. LGMB152) was able to produce six out of seven enzymes analyzed. Several reports have been shown that plant rhizospheric strains exhibit high enzymatic activity, in addition to other antifungal metabolites [48], and the hydrolytic enzymes produced by these strains can degrade the structural matrix of fungal cell walls and therefore act as antifungal factors [49].
LGMB152 play a role on enzyme activity against maize pathogens, and when evaluated the antifungal activity, once more, this strain showed promising results with high activity against five out of the six pathogens evaluated (Alternaria sp., Fusarium verticilioides, Cercosporazeae-maydis, Bipolaris maydis and Diaporthe sp.) and moderate activity against Colletotrichum graminicola. This latest, is an important fungus of maize crops [33,50,51]. However, strain LGMB235, which produced enzymes lipase, chitinase and urease, showed considerable activity only against Alternaria sp. and Cercosporazeae-maydis, suggesting that there was not a direct correlation between the production of extracellular enzymes and phytopathogens inhibition, but enzyme production activity can help to protect plant against pathogenic fungi. Therefore, bio prospecting of PGPB aiming at their use as bio control, has high importance to reduce the use of fertilizers and pesticides favoring a sustainable agriculture.
CONCLUSION
ACKNOWLEDGMENT
REFERENCES
- Ranum P, Peña-Rosas JP, Garcia-Casal MN (2014) Global maize production, utilization, and consumption. Ann N Y Acad Sci 1312: 105-112.
- Cassán F, Maiale S, Masciarelli O, Vidal A, Luna V, et al. (2009) Cadaverine production by Azospirillum brasilense and its possible role in plant growth promotion and osmotic stress mitigation. Eur J Soil Biol 45: 12-19.
- Hungria M, Campo RJ, Souza EM, Pedrosa FO (2010) Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant and Soil 331: 413-425.
- O’Callaghan M (2016) Microbial inoculation of seed for improved crop performance: Issues and opportunities. Appl Microbiol Biotechnol 100: 5729-5746.
- Silva JRC, Souza RM, Zacarone AB, Silva LHCP, Castro MAS (2008) Bactérias endofíticas no controle e inibição in vitro de pseudomonas syringae pv tomato, agente da pinta bacteriana do tomateiro. Ciênc agrotec 32: 1062-1072.
- Assumpção LC, Lacava PT, Dias ACF, Azevedo JL, Menten JOM (2009) Diversidade e potencial biotecnológico da comunidade bacteriana endofítica de sementes de soja. Pesqui Agropec Bras 44: 503-510.
- Ikeda AC, Bassani LL, Adamoski D, Stringari D, Kava Cordeiro V, et al. (2013) Morphological and genetic characterization of endophytic bacteria isolated from roots of different maize genotypes. Microb Ecol 65: 154-160.
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, New York, USA.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697-703.
- Menna P, Hungria M, Barcellos FG, Bangel EV, Hess PN, et al. (2006) Molecular phylogeny based on the 16S rRNA gene of elite rhizobial strains used in Brazilian commercial inoculants. Syst Appl Microbiol 29: 315-332.
- Dowd SC, Zaragoza J, Rodriguez JR, Oliver MJ, Payton PR (2005) Windows .NET network distributed basic local alignment search toolkit (W.ND-BLAST). Bmc Bioinformatics 6: 93.
- Hall TA (2013) Bio Edit 7.2.5. Bio edit: A user-friendly bi ological sequence alignment editor and analysis program for Windows 95/98/NT [http://www.mbio.ncsu.edu/BioEdit/bioedit.html].
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680.
- Tamura K, Dudley J, Nei M, Kumar S (2012) MEGA5: Molecular Evolutionary Genetics Analysis (MEGA) software version 5.0.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A (2012) MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539-542.
- Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160: 47-56.
- Chagas Junior AF, de Oliveira LA, de Oliveira AN, Willerding AL (2010) Capacidade de solubilização de fosfatos e eficiência simbiótica de rizóbios isolados de solos da Amazônia. Acta Sci Agron 32: 359-366.
- Araújo LM, Monteiro RA, Souza EM, Steffens MB, Rigo LU, et al. (2004) GlnB is specifically required for Azospirillum brasilense NifA activity in Escherichia coli. Res Microbiol 155: 491-495.
- Baldani JI, Reis VM, Videira SS, Boddey LH, Baldani VLD (2014) The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: A practical guide for microbiologists. Plant and Soil 384: 413-431.
- Kuss AV, Kuss VV, Lovato T, Flôres ML (2007) Fixação de nitrogênio e produção de ácido indol in vitro por bactérias diazotróficas endofíticas. Pesqui Agropec Bras 42: 1459-1465.
- Ayres M, Ayres Junior M, Ayres DL, Santos AS (2007) Bio Estat. Versão 5.0, Sociedade Civil Mamirauá, MCT - CNPq, Belém, Pará, Brasil.
- Regras para análise de sementes (2009) Ministério da Agricultura, Pecuária e Abastecimento. Regras para análise de sementes, Brasília, Pg no: 399.
- Silva FS, Azevedo CAV (2002) Versão do programa computacional Assistat para o sistema operacional Windows. Rev Bras Prod Agroind 4: 71-78.
- Murugappan RM, Aravinth A, Rajaroobia R, Karthikeyan M, Alamelu MR (2012) Optimization of MM9 medium constituents for enhancement of siderophoregenesis in marine Pseudomonas putida using response surface methodology. Indian J Microbiol 52: 433-441.
- Hankin L, Anagnostakis SL (1975) The use of solid media for detection of enzyme production by fungi. Mycologia 67: 597-607.
- Renwick A, Campbell R, Coe S (1991) Assesment of in vivo screening systems for potential biocontrol agents of Gaeumannomyces graminis. Plant Pathol 40: 524-532.
- Sierra G (1957) A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek 23: 15-22.
- Berg G, Egamberdieva D, Lugtenberg B, Hagemann M (2010) Symbiotic Plant-Microbe interactions: Stress protection, plant growth promotion, and biocontrol by Stenotrophomonas. Symbioses and Stress 18: 445-460.
- Dye DW (1968) A taxonomic study of the genus Erwinia 1. The “amylovora” group. New Zeal J Sci 11: 590-607.
- Noriler SA, Savi DC, Aluizio R, Palácio Cortes AM, Possiede YM, et al. (2018) Bioprospecting and structure of fungal endophyte communities found in the Brazilian biomes, Pantanl, and Cerrado. Front Microbiol 9: 1526.
- Szilagyi-Zecchin VJ, Adamoski D, Gomes RR, Hungria M, Ikeda AC, et al. (2016) Composition of endophytic fungal community associated with leaves of maize cultivated in south Brazilian field. Acta Microbiol Immunol Hung 4: 449-466.
- de Souza R, Beneduzi A, Ambrosini A, da Costa PB, Meyer J, et al. (2013) The effect of plant growth-promoting rhizobacteria on the growth of rice (Oryza sativa L.) cropped in southern Brazilian fields. Plant and Soil 366: 585-603.
- Costa PB, Granada CE, Ambrosini A, Moreira F, Souza R, et al. (2014) A model to explain plant growth promotion traits: A multivariate analysis of 2,211 bacterial isolates. PLoS One 9: 116020.
- Souza R, Ambrosini A, Passaglia LMP (2015) Plant growth-promoting bacteria as inoculants in agricultural soils. Gen Mol Biol 38: 401-419.
- De Bashana LE, Hernandez JP, Bashana Y, Maier R (2010) Bacillus pumilus ES4: Candidate plant growth-promoting bacterium to enhance establishment of plants in mine tailings. Environ Exp Bot 69: 343-352.
- Nautiyal CS, Srivastava S, Chauhan PS, Seem K, Mishra A, et al. (2013) Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol Biochem 66: 1-9.
- Ahemad M, Kibret M (2014) Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J King Saud Uni Sci 26: 1-20.
- Bashan Y, Moreno M, Troyo E (2000) Growth promotion of the seawater-irrigated oilseed halophyte Salicornia bigelovii inoculated with mangrove rhizosphere bacteria and halotolerant Azospirillum spp. Biol Fertil Soils 32: 265-272.
- Enebak SA, Wei G, Kloepper JW (1998) Effects of plant growth-promoting rhizobacteria on loblolly and slash pine seedlings. For Sci 44: 139-144.
- Hernandez JP, de-Bashan LE, Rodriguez DJ, Rodriguez Y, Bashan Y (2009) Growth promotion of the freshwater microalga Chlorella vulgaris by the nitrogen-fixing, plant growth-promoting bacterium Bacillus pumilus from arid zone soils. Eur J Soil Biol 45: 88-93.
- Kloepper JW, Reddy MS, Rodríguez-Kabana R, Kenney DS, Kokolis-Burelle N, et al. (2004) Application for rhizobacteria in transplant production and yield enhancement. Acta Hort 631: 217-229.
- Zahid M, Abbasi MK, Hameed S, Rahim N (2015) Isolation and identification of indigenous plant growth promoting rhizobacteria from Himalayan region of Kashmir and their effect on improving growth and nutrient contents of maize (Zea mays L.). Front Microbiol 6: 207.
- Kuan KB, Othman R, Abdul Rahim K, Shamsuddin ZH (2016) Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilization of maize under greenhouse conditions. PLoS One 11: 0152478.
- Szilagyi Zecchin VJ, Klosowski AC, Ikeda AC, Hungria M, Galli Terasawa LV, et al. (2015) Potential inoculant strains of Brazilian endophytic bacteria for maize (Zea mays L) growth promotion. Int J Agron Agric Res 7: 128-134.
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233-266.
- Araújo FF, Guerreiro RT (2010) Bioprospecção de isolados de Bacillus promotores de crescimento de milho cultivado em solo autoclavado e natural. Ciênc agrotec 34: 837-844.
- Geetha K, Bajithasri AB, Bhadraia B (2014) Isolation of plant growth promoting rhizo bacteria from rizhosphere soils of green gram, biochemical characterization and screening for antifungal activity against pathogenic fungi. Int J Pharm Sci Inv 3: 47-54.
- Ayyadurai N, Ravindra Naik P, Sakthivel N (2007) Functional characterization of antagonistic fluorescent pseudomonads associated with rhizospheric soil of rice (Oryza sativa L.). J Microbiol Biotechnol 17: 919-927.
- Josic D, Ciric A, Sokovic M, Stanojkovic-Sebic A, Pivic R, et al. (2015) Antifungal activities of indigenous plant growth promoting Pseudomonas spp. from alfalfa and clover rhizosphere. Front Life Sci 8: 131-138.
- Duncan KE, Howard RJ (2010) Biology of maize kernel infection by Fusarium verticillioides. Mol Plant Microbe Interact 23: 6-16.
- Neves DL, Silva CN, Pereira CB, Campos HD, Tessmann DJ (2015) Cercospora zeina is the main species causing gray leaf spot in southern and central Brazilian maize regions. Trop Plant Pathol 40: 368-374.
Citation: Ikeda AC, Zecchin VJS, Savi DC, Kava V, Glienke C, et al. (2018) Bio Prospecting Plant Growth-Promoting Bacteria Isolated from Maize (zea mays l.) Roots. J Biotech Res Biochem 1: 003
Copyright: © 2018 Angela Cristina Ikeda, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

