Cloning, Expression, and Purification of the rE2 Protein Human Papillomavirus type 16
*Corresponding Author(s):
Saucedo Mendiola María LeticiaFacultad De Ciencias Quimicas, UJED, Durango, Mexico
Tel:+618 1380439,
Email:sauced101@yahoo.com.mx
Abstract
Human papillomavirus (HPV) infection is the most common sexually transmitted disease, both in men and women, and the persistence of this infection is the main causative agent of cervical-uterine cancer. HPV 16 protein E2 is important in viral replication and transcription. For this reason, it is of utmost importance to have a reliable method of expression and purification of recombinant E2 (rE2) proteins to carry out experiments focused on the development of drugs against HPV. The main goal of this work was to obtain the HVP purified rE2 protein using the pET28a (+) Vector Systems. E2 gene amplification was performed by end-point PCR and later was cloned in the vector pGEM-t- easy in the E.Coli TOP 10F 'strain, the E2 amplicon was digested, purified and subcloned in the expression vector pET -28a (+) in the E. coli BLR21 strain, it was cultivated in LB medium added with kanamycin, 0.5mM IPTG was used as inducer and it was cultivated for 10 hours at 37 ° C/200rpm additional. The 39 kDa weight rE2 protein was purified using affinity chromatography. SDS-PAGE 10% analysis of protein purification showed a band corresponding to 39 kDa of molecular weight corresponding to the rE2.
Keywords
E2 protein; Expression, HPV
INTRODUCTION
Cervical-uterine cancer (CaCu) is the 3rd most common cancer among women worldwide, with an estimated 569,847 new cases and 311,365 deaths in 2018 [1]. CaCu is the second leading cause of cancer death in Mexican women, its main etiological agent being HPV [2]. Among the different types of virus, type 16 is the most prevalent worldwide [3]. Currently, there is no antiviral drug available despite the high incidence of these viruses in the sexually active population; therefore, the need for antiviral agents to treat HPV-associated diseases remains relevant [4].
All HPV are non-enveloped double stranded DNA viruses. Their genomes are circular and approximately 8 kilo base-pairs in size. Most encode eight major proteins, 6 located in the “early” region 2 in the “late” region [5]. The “early” proteins are regulatory in function. For example, they play roles in HPV genome replication and transcription, cell cycle [6]. The late region is composed of the proteins L1 and L2, which comprise the capsid of the virus necessary for the transmission of the virus [7].
The HPV16 protein E2 is an attractive target for the development of anti-HPV agents because of its direct involvement in viral DNA replication and transcription and in the segregation of the viral genome during cell division. As a replication initiation factor, E2 binds with high affinity to specific sites located within the viral origin (ori) to help recruit it to the E1 helicase [8,9].
The E2 gene is one of the most conserved genes and is made up of approximately 1089 base-pairs (bp), it encodes a 39 KiloDaltons (kDa) nuclear protein, which is divided into three functional domains. The first, at the amino-terminal end, the second domain is the hinge or central domain and the third domain is the carboxyl-terminal end, it is dimerizing and binding to DNA, of approximately 100 amino acids. The E2 protein of all viral strains has in common the fact that they bind to a palindromic DNA sequence (ACCgNNNNcGGT, the small letters indicate the preferred nucleotides; the NNNN region is called the spacer), referred to as the E2 binding site [9,10].
MATERIAL AND METHODS
Amplification of HPV-16 E2 Gene
Genomic DNA was obteined from a biological sample with HPV-16 and used as template to amplify the E2 gene with the primers forward and reverse (forward primer: 5'- GGATCCAGACTCTTTGCC-3'; reverse primer: 5'-TACTGGATTTATGTCTATATGACAAAGCTT-3') containing BamHI and HindIII restriction enzyme sites, respectively, designed with oligoanalyzer tool from IDT's www.idtdna.com/calc/analyzer. The PCR assay was performed in a final volume of 25 μL, contained 0.4 µL of genomic DNA, 5 µL of 5X buffer, 0.5 µL of dNTP`s, 0.5 µL of the direct oligonucleotide, 0.5 µL of the reverse oligonucleotide, 0.1 µL Taq polymerase and nuclease-free water. The amplification reaction was carried out in a thermal cycler (Sure Cycler 8800) with denaturation at 94°C for 4 min follow by 30 amplification cycles of denaturation, 51.5°C for 1 min, 72°C for 1 min. The E2 amplicons were electrophoresed on 1% agarose gels, containing 0.1% ethidium bromide, (GABrET) and visualized on a Bio-Rad ChemiDoc [11] photo documenter and stored at -4°C until use.
Plasmid construction E2- pGEM-t-easy
PCR products were purified using the Wizard® Sv Gel and PCR clean-up system kit [12] (Promega Corporation 2800 Woods Hollow Road Madison, WI 53711 USA), then digested with BamHI [13] and HindIII [14] (Promega) and ligated in plasmid pGEM-t-easy (Promega), 0.5 ng/µL ligation product were used to transform E.Coli TOP10F ' calcium-competent cells [15].
Subcloning of the HPV-16 E2 amplicon into expression vector pET-28a (+)
The plasmid pGEM-t-easy-E2 was digested with the enzymes HindIII and BamHI and purified using the Wizard Sv Gel and PCR clean-up system kit.
HPV-16 E2 DNA fragment was subcloned in His6 tag sequence at the N-terminus of pET-28a (+) vector (Novagen) using HindIII and BamHI restriction sites. E2- pET-28a (+) was transformed into E. coli DH5-alpha calcium- competent cells by heat shock [15].
Expression of recombinant HPV- E2
E2-pET-28a (+) was introduced into E. coli BL21 (DE3) calcium- competent cells by heat shock and the insert was confirmed by DNA sequencing (Institute of Biotechnology of the UNAM).
E. coli pre-cultures of the strain harboring the plasmids were grown overnight on Luria–Bertani (LB) broth (1% peptone, 1.0% NaCl and 0.5% yeast extract) containing 50 μg/mL Gibco® Kanamycin at 37°C and 200 rpm.
The expression of His6-E2 was induced by the addition of isothiogalactopyranoside (IPTG) using concentrations of 0.25 mM and 0.5 mM. Briefly, 200 mL of Lb broth containing 50 μg/mL kanamicin was inoculated with 1% (v/v) of overnight culture and incubated at 37°C at 200 rpm until the OD600 reaches 0.5, IPTG was then added and the induction continued at 37°C for 10 h. Cells were pelleted by centrifugation and resuspended in buffer Tris/NaCl 50 mM, pH 8.0 .To the resulting cell suspension, a lysis buffer was added (Lysozyme 100 mg/mL, Tris/NaCl 50 mM pH 8.0) and incubated overnight at 37°C/200 rpm. The lysate was centrifuged at 4000 rpm for 20 min and the supernatant was separated.
The presence of the expressed protein of interest was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of an aliquot of the supernatant.
Purification of recombinant E2
The recombinant protein was purified by TALON metal affinity resin (Clontech, USA) [16] as per supplier’s instruction. 1 mL of the resin was poured into a column and equilibrated with equilibrium buffer (containing 10 mM imidazole, pH 7.4), After being washed with buffer (14.94 mL of HisTALON equilibrium buffer mixed with 1.06 mL of HisTALON elution buffer), the bound protein was eluted with HisTALON elution buffer (containing 150 mM imidazole) and fractions of 1mL were collected in Eppendorf tubes. The CO+2-NTA purification fractions were analyzed on 10% SDS–PAGE gels stained with Coomassie Blue (Bio-SafeTM Coomassie G-250 Stain) and protein concentrations were measured on the Thermo Fisher Scientific NanoDropTM at 280 nm.
RESULTS
Amplification of the HPV-16 E2 gene
A fragment of 1089 bp was generated by PCR amplification of genomic DNA of HPV-16 corresponding to the size in bp of the E2 (Figure 1). Following digestion of the PCR product with BamH1 and HindIII, the fragment was cloned between BamH1 and HindIII site of pGEM-T-easy to generate the plasmid E2-pGEM-T-easy.
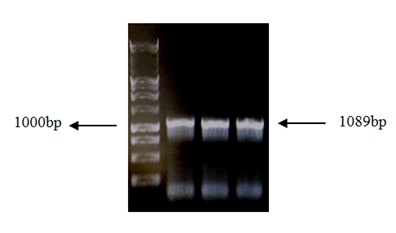 Figure 1: Electrophoresis of amplicon E2 in 1% GaBrEt. Lane 1: Invitrogen 1Kb Plus DNA Ladder™, lanes 2, 3 and 4: Single expected band of purified E2 amplicon (1089 bp).
Figure 1: Electrophoresis of amplicon E2 in 1% GaBrEt. Lane 1: Invitrogen 1Kb Plus DNA Ladder™, lanes 2, 3 and 4: Single expected band of purified E2 amplicon (1089 bp).
The plasmid E2-pGEM-t-easy was digested with the enzymes HindIII and BamHI (Figure 2), and HPV16 E2 DNA fragment was subcloned in His6 tag sequence at the N-terminus of pET-28a (+) vector (Figure 3).
The constructs in the E. coli BL21 (DE3) were stored in eppendorf tubes with LB with Glycerol 10% and frozen at -80°C.
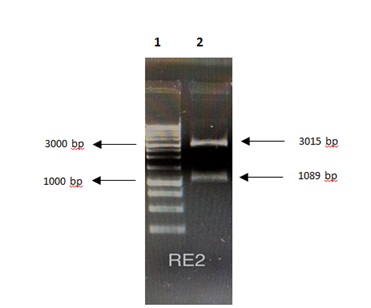 Figure 2: Electrophoresis in GaBrEt 1% of the digested plasmid pGEM-T-easy-E2. Lane 1: Invitrogen 1 Kb Plus DNA Ladder ™, lane 2: 3015bp pGEM-t-easy vector, and 1089bp E1 gene.
Figure 2: Electrophoresis in GaBrEt 1% of the digested plasmid pGEM-T-easy-E2. Lane 1: Invitrogen 1 Kb Plus DNA Ladder ™, lane 2: 3015bp pGEM-t-easy vector, and 1089bp E1 gene.
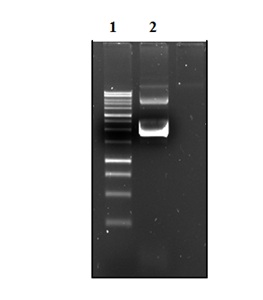 Figure 3: Electrophoresis in GaBrEt 1% of plasmid DNA pET-28a (+) / E2. Lane 1: Invitrogen 10 Kb Plus DNA Ladder ™. Lane 2: plasmid DNA pET-28a (+) / E2.
Figure 3: Electrophoresis in GaBrEt 1% of plasmid DNA pET-28a (+) / E2. Lane 1: Invitrogen 10 Kb Plus DNA Ladder ™. Lane 2: plasmid DNA pET-28a (+) / E2.
Successful cloning of the gene was confirmed by restriction endonuclease analysis (Figure 4) and sequencing. A sequence alignment of HPV-E2 against the PaVe database (pave.niaid.nih.gov) using GGSEARCH/GLSEARCH revealed a high degree of homology (98.6%) (Figure 5).
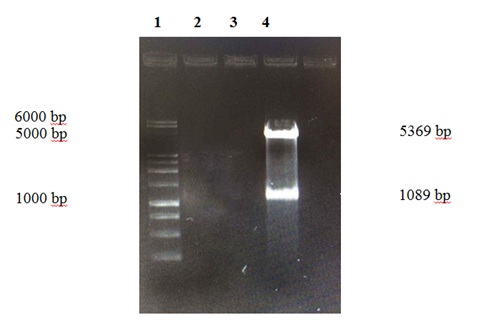 Figure 4: Electrophoresis in GaBrEt 1% of the digestion of plasmid pET-28a (+) - E2 Lane 1. Invitrogen 10 Kb Plus DNA Ladder ™ Lane 4. Vector pET-28a (+) 5.369bp and E2 gene of 1.08
Figure 4: Electrophoresis in GaBrEt 1% of the digestion of plasmid pET-28a (+) - E2 Lane 1. Invitrogen 10 Kb Plus DNA Ladder ™ Lane 4. Vector pET-28a (+) 5.369bp and E2 gene of 1.08

Figure 5: Alignment of HPV-16 E2 sequenced and the E2 sequence reported in PaVE
Overexpression and purification of recombinant HPV-E2
Induction of protein expression was carried out at concentrations of 0.25 and 0.5 mM IPTG; higher expression was observed at 0.5 mM. The figure 6 show the analysis of cell extracts after induction with 0.5mM IPTG in 10% SDS-PAGE. The recombinant protein was purified using affinity chromatography and resulted in pure protein according to SDS-PAGE analysis. The molecular mass of the purified protein 39 kDa based was based on its mobility in SDS-PAGE, this value agreed with the molecular size predicted from its amino acid composition (Figure 7), the yield was 1 mg of protein/liter of culture.
The purified rE2 protein was stored in 1.5 ml eppendorf tubes at 7°C. The protein showed good stability for up to 4 months at storage temperature.
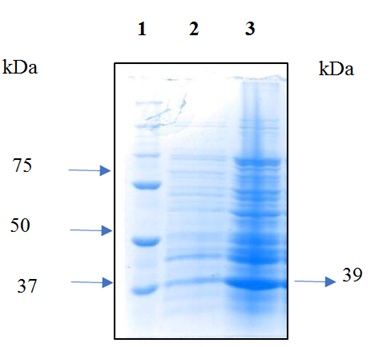 Figure 6: Analysis of cell extracts after induction in 10% SDS-PAGE. Lane 1: 10-250kDa protein molecular weight marker The Thermo Scientific™. Lane 2: negative control. Lane 3: induction with 0.5 mM IPTG at 37°C for 10 h.
Figure 6: Analysis of cell extracts after induction in 10% SDS-PAGE. Lane 1: 10-250kDa protein molecular weight marker The Thermo Scientific™. Lane 2: negative control. Lane 3: induction with 0.5 mM IPTG at 37°C for 10 h.
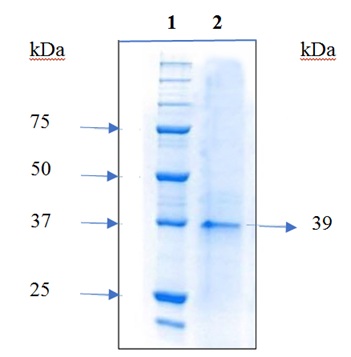
Figure 7: 10% SDS-PAGE of purified HPV16 rE2 protein. Lane 1: Molecular weight marker of proteins from 10-250 kDa The Thermo Scientific™. Lane 2: Purified 39 kDa rE2 protein.
DISCUSSION
Currently, there is no antiviral drug available for the treatment of HPV-associated diseases despite the high incidence of these viruses in the sexually active population. Therefore, the need for antiviral agents to treat HPV associated diseases remains relevant [17]. HPV DNA replication is an attractive target for the development of HPV antivirals since inhibition of HPV DNA replication would result in fewer genomes available for viral protein synthesis, as well as fewer genomes for HPV. Integration a critical step in the development of HPV-dependent cancers.
During the analysis of the open reading frame of the E2 gene as well as of the HPV16 sequence for the design of primers, 50% and 60% of guanines and cytokines were detected respectively, which give these sequences great stability.
The expression vector used in this work was pET 28a (+), in which the protein is expressed as a fusion protein with a histidine tail at its N-terminal end. The pET vectors that have the T7 promoter are frequently used for the expression of recombinant proteins, as they produce high levels of transcription in E. coli [18].
There are some studies in which the same vector has been used for the expression of the E2 protein of different types of HPV and inducing with IPTG at different concentrations from 0.3 to 1 mM with incubation temperatures of 16-30 °C [19-23].
It was observed that the time after induction depended on the IPTG concentration and incubation temperatures, thus when temperatures were low, the incubation time increased as shown in the work [23]. Where the incubation temperature was of 16 ° C, obtaining the expression, until 16 h after induction, in contrast to those found in this investigation where the expression of E2 was induced with 0.5 mM IPTG and the incubation time was reduced to 10 h after induction, from that time on, the expression was no longer increased.
CONCLUSION
Based on the results of the sequencing of the plasmid pET28a (+)/E2 (98.6%) and the molecular weight (39 kDa) observed in the SDS-PAGE analysis, the protein obtained is the rE2 of HPV16.
REFERENCES
- https://www.hpvcentre.net/statistics/reports/XWX.pdf
- http://www.beta.inegi.org.mx/contenidos/saladeprensa/aproposito/2018/cancer2018_Nal.pdf
- García J, Molina J, Blasco B (2010) The human papillomavirus and cervical cancer. A review of the updated history of cervical cancer research in Venezuela. Invest Clín. 51: 193-208.
- Álvarez AA, Sepúlveda A, Siller LF (2012) Carcinogénesis inducida por el virus del papiloma humano. Investigaciones Andinas. 14: 438-456.
- Graham SV (2010) Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future microbiology 5: 1493-1506.
- Tang S, Tao M, McCoy JP, Zheng ZM (2006) The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol 80: 4249-4263.
- Graham SV (2006) Late events in the life cycle of human papillomaviruses. Papillomavirus research: from natural history to vaccines and beyond.Caister Academic Press; Wymondham, Norfolk 193-212.
- WangY, Coulombe R, Cameron DR, Thauvette L, Fink D, et al. (2004) Crystal structure of the E2 transactivation domain of human papillomavirus type 11 bound to a protein interaction inhibitor. J Biol Chem 279: 6976-6985.
- Rashmi SH, Androphy EJ (1998) Crystal Structure of the E2 DNA-binding Domain from Human Papillomavirus Type 16: Implications for its DNA Binding-site Selection Mechanism. J Mol Biol 284: 1479-1489.
- Hegde RS (2002) The papillomavirus E2 proteins: Structure, function, and biology. Annu Rev Biomol Struct 31: 343-360.
- https://www.bio-rad.com/en-uk/category/chemidoc-imaging-systems?ID=NINJ0Z15
- https://www.promega.in/resources/protocols/technical-bulletins/101/wizard-sv-gel-and-pcr-cleanup-system-protocol/
- https://worldwide.promega.com/products/cloning-and-dna-markers/restriction-enzymes/bamhi/?catNum=R6021
- https://www.promega.in/products/cloning-and-dna-markers/restriction-enzymes/hindiii/?catNum=R6041
- Sambrook J, Russell WD (2006) Preparation and Transformation of Competent E. coli Using Calcium Chloride. Cold Spring Harb Protocols.
- https://www.takarabio.com/assets/documents/User%20Manual/TALON%20Metal%20Affinity%20Resins%20User%20Manual%20(PT1320-1)_031716.pdf
- Fradet-Turcotte A, Archambault J (2007) Recent advances in the search for antiviral agents against human papillomaviruses. Antivir Ther 12: 431-451.
- Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: Advances and challenges. Front Microbiol 5: 1-17.
- Sedman T, Stenlund A (1997) Binding of the E1 and E2 Proteins to the Origin of Replication of Bovine Papillomavirus. Jurnal and Virology 55: 2887-2896.
- Chen G, Stenlund A (2002) The E1 initiator recognizes multiple overlapping sites in the papillomavirus origin of DNA replication. Jurnal of Virology 75: 292-302.
- White PW, Titolo S, Brault K, Thauvette L, Pelletier A, et al. (2003) Inhibition of Human Papillomavirus DNA Replication by Small Molecule Antagonists of the E1-E2 Protein Interaction. The journal of biological chemistry 201: 26765-26772.
- Hernandez REE, Burns J, Zhang E, Walker HF, AllenS, et al. (2008) Dimerization of the Human Papillomavirus Type 16 E2 N Terminus Results in DNA Looping within the Upstream Regulatory Region. Journal of Virology 251: 4853-4861.
- Kantang W, Chunsrivirot S, Muangsin N, Poovorawan Y, Krusong K, et al. (2016) Design of peptides as inhibitors of human papillomavirus 16 transcriptional regulator E1-E2. Chem Biol Drug Des 88: 475-484.
Citation: Alonso GMJ, Estela FZM, Ismael LA, Veronica LC, Luis RBJ, et al.(2020) Cloning, expression, and purification of the rE2 protein Human Papillomavirus type 16. J Protein Res Bioinform 2: 012
Copyright: © 2020 Gándara Mireles Jesús Alonso, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.