Comparative Analysis of Amino acid Sequence Diversity and Physiochemical Properties of Peroxidase Superfamily
*Corresponding Author(s):
Shamsher S KanwarDepartment Of Biotechnology, Himachal Pradesh University, India
Tel:+91 8219994569,
Email:kanwarss2000@yahoo.com
Abstract
Peroxidases superfamily is amongst the most widespread enzyme among living organisms, which maintains vital role(s) in many biological processes. Found in variety of species ranging from microbe to animals their main functions differ from organism to organism. Peroxidases mainly catalyze the oxidation of diverse substrates using hydrogen peroxide (H2O2). Peroxidases are a big family of enzymes which comprises of animal peroxidase and plant peroxidase superfamily. Plant peroxidases are further divided into three classes on basis of their origin and functions. In the present study an attempt has been made to relate three classes of plant peroxidase superfamily on the basis of enzyme’s amino acid sequences and physiochemical properties by using bioinformatics tools (Assistat version-7.7, Clustal W, etc). Further phylogenetic relation was set up among them after doing multiple sequence alignment. Significant difference in the total number of amino acid residues among the classes was sufficient to determine factors like stability and nature (charge) of the enzyme. It was observed that class III peroxidase appeared to be the most stable class in plant peroxidase superfamily while class I being the least stable one. The study provides more insight about how closely the peroxidase classes are related to each other structurally. Distribution of amino acids also helped in stating the intracellular nature of class I plant peroxidase which differs from rest two classes of the plant peroxidase family which are extracellular in nature.The study resulted in giving a conclusive idea about the functional group of different classes of peroxidase and specific characters of all three families. This dry lab study was done using the amino acid sequences of 5 organisms of each class of plant peroxidase superfamily, retrieved from NCBI (http://www.ncbi.nlm.nih.gov/protein) and UniProt proteomic server (http://www.expasy.org). The choice of organisms for each class was based upon their ability produce peroxidase in laboratory conditions with a high yield factor.
Keywords
Amino acids, Peroxidase, Physiochemical Properties
INTRODUCTION
Peroxidases widely present in nature, acts as a key natural antioxidant. Peroxidaseacts as an oxidizing agent,and facilitate in decomposition aided with decomposition of H2O2 [1]. They have different roles in different organisms as in mammals they help in immune system functions or in hormone regulations.In plants, they are implicated in metabolism of auxin, lignin, super besides cell wall formation and defense against pathogens. Plant peroxidases (peroxidase EC: 1.11.1.7) are haem-proteins, which help in hydrogen peroxide (H2O2) mediated catalysis [2]. Peroxidases also play important roles in protection of plant leaves from salt-induced oxidative damage [3]. By the early 1900s, as yet unknown enzyme at work in human body were labeled as “catalases” while the simultaneous observation that plants and animals utilized polyphenols to degrade H2O2 lead to the term “peroxidases” [4]. More than 30 different kinds of peroxidases are found in humans whereas Arabidopsis thaliana has as many as 130 different peroxidases [5]. Peroxidases from protists, fungi and prokaryotic organisms are known to promote virulence [6]. Although, the most well-known and best studied peroxidase is horseradish peroxidase [7] yet bacterial peroxidases have attracted comparatively more attention than plant peroxidases [8].
In peroxidase, the bound cofactor necessary for its activity is heme [4]. Most heme peroxidases belong to two superfamilies at molecular level, one present in plants, bacteria and fungi [9,10], and the other-one found in animals [11,12]. These have iron (III) protoporphyrin IX (ferriprotoprophyrin IX) as the prosthetic group and heme is a complex between an iron ion and a molecule protoporphyrin IX [13,14]. Catalytic cycle involves distinct intermediate enzyme forms [15].
For most of the peroxidases, the optimal substrate is H2O2 but others are much active with organic hyper oxides such as lipid oxides. Peroxidases are used in clinical immunoassay and enzyme biochemistry and for the colorimetric measurement of biological materials [16]. Some novel applications of peroxidases include waste-water (containing phenolic compounds) treatment, removal of peroxide from materials such as industrial wastes and foodstuffs and synthesis of different aromatic chemicals [17]. Peroxidases have potential for bio-remediation of wastewater contaminated with cresols, chlorinated and non-chlorinated phenols, and for bio-pulping, bio-bleaching in paper industry and textile-dye degradation [18].
Based on differences in primary structure, the peroxidase superfamily can be further divided into three classes I, II and III. Class I peroxidases are intracellular in nature, present in plants, bacteria and yeast such as microbial cytochrome peroxidase (EC1.11.1.5), bacterial catalase-peroxidase (EC1.11.1.6) and ascorbate (EC1.11.1.11) peroxidase. Class II peroxidases, are fungal peroxidases exclusively, thathave major role in lignin biodegradation [19]. They include lignin peroxidase (LiPs; EC1.11.1.14) and Mn2+ dependent peroxidase (MnPs: EC1.11.1.13). Versatile peroxidases (VP; EC 1.11.1.16), have a molecular structure that is hybrid between MnPs and LiPs [20] and may result in catalysis of non-phenolic and phenolic substrates like LiPs [21]. Class III peroxidases (EC 1.11.1.7), widely distributed in plant kingdom [22], include soybean peroxidase (SBP), horseradish peroxidases (HRP), peanut peroxidase (PNP), etc. play a vital role in the life cycle of many plants [23]. In plants class III peroxidases are involved in functions like cell wall metabolism [24], lignification [25], suberization [26], auxins metabolism [27], wound healing [28], release of reactive oxygen species (ROS), reactive nitrogen species (RNS) metabolism [29], fruit growth and ripening [30] and defense against pathogens [31]. Plant peroxidase superfamily is of a high economic value due to its diverse applications [32]. Though a large no of data is present for plant peroxidase superfamily, never an attempt was made to compare and inter-relate different classes of this superfamily. Due to high economic importance, finding the relation and difference among them can help us to understand the nature better which can result in enhancing their economic value. In the present study an attempt has been made to analyze three classes of plant peroxidase superfamily using different bioinformatics tools.
MATERIALS AND METHODS
Retrieval of amino acid sequences
The amino acid sequences of peroxidases for different classes were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/protein), UniProt proteomic server (http://www.expasy.org/) shown in table 1. Organisms were selected on the basis of their ability to produce peroxidase in laboratory conditions with a high yield factor. Total five organisms were selected for each class so that the data has a high value of significance upon computation.
|
Sr. No |
Microorganisms |
Accession number |
|
Class I peroxidases |
||
|
1. |
Helicobacter pylori B8 |
CBI65628.1 |
|
2. |
Escherichia coli str. K-12 |
YP_491917.1 |
|
3. |
Rhodopirellula baltica SH 1 |
NP_863829.1 |
|
4. |
Alcanivorax borkumensis SK2 |
YP_694094.1 |
|
5. |
Campylobacter coli 15-537360 |
AGZ20833.1 |
|
Class II peroxidases |
||
|
1. |
Ceriporiopsis subvermispora B |
EMD32649.1 |
|
2. |
Fomitopsis pinicola FP-58527 SS1 |
EPT05268.1 |
|
3. |
Ganoder malucidum |
ACA48488.1 |
|
4. |
Schizophyllum sp. F17 |
AGO86670.2 |
|
5. |
Cerrena unicolor |
AGS19356.1 |
|
Class III peroxidases |
||
|
1. |
Gossypium hirsutum |
ACJ11766.1 |
|
2. |
Beta vulgaris |
CAK22416.1 |
|
3. |
Solanum lycopersicum |
CAG25463.1 |
|
4. |
Stylosanthes humilis |
AAB67737.1 |
|
5. |
Nelumbo nucifera |
ABN46984.1 |
Table 1: Different organisms producing plant enzymes.
Analysis of some important physiochemical parameters
The physiochemical data of amino acid sequences were generated from the SwissProt and Expert Protein Analysis System (ExPASy). The data for various parameters was generated by using online tool ProtParam and Compute pI/Mw at the ExPASy. The molecular weight (kDa) of sequences was calculated byaverage isotopic masses addition to amino acids in the protein and then deducing average isotopic mass of one molecule of water. Isoelectic point (pI) was calculated using pKa values of amino acids according [33].
Primary structure analysis
The amino acid compositions of proteases was calculated from the ProtParam tool which is available on ExPASy. Molar extinction coefficient of protease proteins was calculated according to equation; E(Prot) = Numb (Tyr)* Ext (Tyr) + Numb (Trp)* Ext (Trp) + Numb (Cystine)* Ext (Cystine)
Aliphatic index of various sequences was obtained using the formula:
Aliphatic index = X (Ala) + a * X (Val) + b * [X (Leu) + X (Ile)] [34]
Where, X (Ile), X (Val), X (Ala), and X (Leu) are mole percent (100 X mole fraction) of isoleucine, valine, alanine, and leucine. The coefficient a and b are the relative volume of valine side chain and of leu/Ile side chain to the side chain of alanine, with the help of ProtParam tool [34].
Secondary structure analysis
Secondary structure analysis included the analysis of number of β-turn, α-helices, β-sheet and extended strand performed by ExPASy SIB Bioinformatics SOPMA tool (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page = npsa_sopma.html) [35]. To utilize this tool sequences were submitted in FASTA format [36].
Tertiary structure analysis
PHYRE2 software (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) was used to build the 3D models of different peroxidases and their validation was done using the Ramachandran plot constructed using MOL probity (http://molprobity.biochem.duke.edu/) [37].
Multiple sequence alignment and phylogenetic analysis
The phylogeny of three classes of peroxidases was generated followed by alignment using MEGA-X tool which is the standalone tool for the multiple sequence alignments and phylogenetic analysis [38].
RESULTS AND DISCUSSION
Computational analysis of some important physiochemical parameters and amino acids of peroxidases
The present study reports a detailed comparison of various physiochemical parameters and amino acids among the three classes of plant peroxidases. The result of analysis of physiochemical properties and amino acids content has been tabulated in table 1 & 2. Overall analysis revealed theoretical PI, total number of positively charged residues (Arg + Lys), extinction coefficient, instability index and negative charged residues (Asp + Glu) to be statistically significant among all the groups of serine proteases (Table 1).The total number of amino acid residues in the three classes of peroxidases differed substantially as the number of aminoacid was found to be in the range of 341.0 to 465.0 for class I peroxidases, 332.0 to 364.0 for class II peroxidases and 319.0 to 331.0 for class III peroxidases. Similarly, molecular weight range was 36861.2 to 51570.5 Dalton for class I peroxidases, 34825.4 to 38643.3 Dalton for class II peroxidases and 33781.1 to 37528.1 Dalton for class III peroxidases. On the other hand, theoretical pI ranged from 4.72 to 9.06 for class I peroxidases, 4.12 to 5.33 for class II peroxidases, 5.38 to 8.98 for class III peroxidases, which was found to be statistically significant. The pI value below 7 indicates that a protein is acidic while above 7 indicates a protein to have basic structure. The total counts for the negatively charged (Asp + Glu) and positively charged residues (Arg + Lys) were found to be statistically significant and higher in case of class I peroxidases. Extinction coefficient was found to be statistically significant which ranged from 27180.0 to 68675.0 for class I peroxidases, 6000.0 to 21470.0 for class II peroxidases, 13575.0 to 23420.0 for class III peroxidases and higher in case of the class I peroxidases. Other significant physiochemical parameter i.e. instability index was also found to be statically significant and ranged between 24.13 to 52.14 for class I peroxidases, 42.18 to 55.71 for class II peroxidases, 29.12 to 42.62 class III peroxidases. Higher the aliphatic index more is the stability of a given compound. A negative value for GRAVY indicates that a protein is non-polar and thus has a greater interaction with water [36].
Parameters |
Microorganisms |
1 |
2 |
3 |
4 |
5 |
|
Number of amino acids
|
Class I |
350.0 |
465.0 |
424.0 |
402.0 |
341.0 |
| Class II | 364.0
|
332.0 |
364.0 |
359.0 |
360.0 |
|
| Class III | 323.0 | 326.0 | 332.0 | 319.0 | 331.0 | |
| Molecular weight (Dalton)
|
Class I |
38815.1
|
51570.5
|
46289.7
|
44236.0
|
36861.2
|
| Class II | 38227.9
|
34825.4
|
38110.5
|
37829.4
|
37678.3
|
|
| Class III | 35729.6
|
35499.1
|
35856.7
|
33781.1
|
37528.1
|
|
| Theoretical pI
|
Class I |
9.06 | 5.98 | 4.72 | 4.87 | 8.16 |
| Class II | 4.12 | 5.33 | 4.42 | 4.78 | 4.47
|
|
| Class III | 5.38
|
8.98
|
7.55
|
7.06
|
8.44
|
|
| Total number of negatively charged residues (Asp + Glu)
|
Class I |
37.0 |
58.0 |
63.0 |
48.0 |
39.0 |
| Class II | 44.0
|
34.0
|
40.0 |
43.0 |
46.0 |
|
| Class III | 39.0
|
32.0
|
27.0
|
27.0
|
44.0 |
|
| Total number of positively charged residues(Arg + Lys)
|
Class I |
46.0 |
52.0 |
36.0 |
28.0 |
41.0 |
| Class II |
14.0 |
25.0 |
17.0 |
24.0 |
22.0 |
|
| Class III | 34.0
|
41.0 |
28.0 |
27.0
|
48.0
|
|
| Extinction coefficient(M-1cm-1) at 280nm
|
Class I |
31775.0 | 68675.0 | 27765.0 | 43110.0 | 27180.0 |
| Class II | 11500.0
|
17210.0
|
6000.0
|
21470.0
|
6000.0
|
|
| Class III | 13575.0 | 16430.0 | 15065.0 | 23420.0 | 17460.0 | |
| Instability index
|
Class I |
31.19
|
38.93
|
41.04
|
52.14
|
24.13
|
| Class II | 55.71
|
48.71
|
43.72
|
42.50
|
43.55
|
|
| Class III | 36.48 | 29.12 | 36.89 | 41.48 | 42.62 | |
| Aliphatic index |
Class I |
83.54
|
80.19
|
69.62
|
71.14
|
86.72
|
| Class II | 83.96
|
83.61
|
78.32
|
80.78
|
85.42
|
|
| Class III | 75.82
|
92.33
|
85.69
|
83.67
|
84.53
|
|
| Grand average of hydropathicity (GRAVY)
|
Class I |
-0.234
|
-0.395
|
-0.438
|
-0.433
|
-0.240
|
| Class II | 0.112
|
-0.027
|
0.060
|
-0.035
|
0.025
|
|
| Class III | -0.167
|
-0.095
|
0.026
|
-0.082
|
-0.362
|
Table 2: Physiochemical parameters of various microorganisms calculated using ProtParam tool at ExPASy proteomic server.
Class I peroxidase: 1. Helicobacter pylori B8 2. Escherichia coli str. K-12 3. Rhodopirellula baltica SH1 4. Alcanivorax borkumensis SK2 5.Campylobacter coli 15-537360
Class II peroxidase: 1. Ceriporiopsis subvermispora B 2. Fomitopsis pinicolaFP58527SS1 3. Ganoder malucidum 4. Schizophyllum sp. F17 5. Cerrena unicolor
Class III peroxidase: 1. Gossypium hirsutum 2. Beta vulgaris 3. Solanum lycopersicum 4. Stylosanthes humilis 5. Nelumbo nucifera
Primary structure analysis
The comparison of individual amino acid composition has revealed that in case of class I peroxidases Glu (E), Lys (K), Trp (W) and Tyr (Y) were found to be predominantly higher. Class II peroxidases revealed the presence of Ala (A), Asp (D), Gln (Q), Gly (G), His (H), Ile (I), Phe (F), Pro (P) and Thr (T) to be higher in comparison to other two classes of peroxidases. On the other hand Arg (R), Asn (N), Cys (C), Leu (L), Met (M), Ser (S) and Val (V) to be predominantly higher in case of the class III peroxidases (Table 3).
|
Amino acid |
Peroxidase class |
1 |
2 |
3 |
4 |
5 |
|
|
Ala (A) |
Class I peroxidases |
5.4 |
9.9 |
11.1 |
7.5 |
9.7 |
|
|
Class II peroxidases |
12.1 |
14.5 |
11.8 |
12 |
11.9 |
||
|
Class III peroxidases |
9 |
5.5 |
6.3 |
6 |
7.6 |
||
|
Arg (R) |
Class I peroxidases |
2.6 |
4.1 |
4 |
4.5 |
1.5 |
|
|
Class II peroxidases |
3.3 |
4.5 |
2.7 |
3.3 |
2.5 |
||
|
Class III peroxidases |
5.3 |
3.4 |
5.1 |
4.1 |
7.3 |
||
|
Asn (N) |
Class I peroxidases |
4.6 |
4.1 |
5.7 |
6.7 |
6.2 |
|
|
Class II peroxidases |
3.3 |
4.2 |
4.5 |
3.6 |
2.8 |
||
|
Class III peroxidases |
6.8 |
5.5 |
5.1 |
5.3 |
4.5 |
||
|
Asp (D) |
Class I peroxidases |
5.1 |
6.7 |
6.4 |
6 |
4.4 |
|
|
Class II peroxidases |
6.3 |
6.6 |
7.2 |
7.1 |
8.3 |
||
|
Class III peroxidases |
7.7 |
5.2 |
5.4 |
6.6 |
8.2 |
||
|
Cys (C) |
Class I peroxidases |
1.7 |
1.5 |
1.4 |
1.2 |
1.2 |
|
|
Class II peroxidases |
2.2 |
1.2 |
2.2 |
2.2 |
2.2 |
||
|
Class III peroxidases |
2.5 |
3.1 |
2.4 |
2.8 |
3 |
||
|
Gln (Q) |
Class I peroxidases |
3.1 |
4.9 |
3.3 |
4 |
2.6 |
|
|
Class II peroxidases |
5.8 |
3.3 |
3.6 |
4.7 |
4.7 |
||
|
Class III peroxidases |
3.4 |
2.5 |
4.5 |
3.8 |
2.4 |
||
|
Glu (E) |
Class I peroxidases |
5.4 |
5.8 |
8.5 |
6 |
7 |
|
|
Class II peroxidases |
5.8 |
3.6 |
4.7 |
4.7 |
4.4 |
||
|
Class III peroxidases |
4.3 |
4.6 |
2.7 |
1.9 |
5.1 |
||
|
Gly (G) |
Class I peroxidases |
8.3 |
7.7 |
8.3 |
7.7 |
8.5 |
|
|
Class II peroxidases |
8.5 |
8.4 |
8.5 |
8.6 |
8.6 |
||
|
Class III peroxidases |
6.8 |
8.9 |
9.6 |
10.7 |
4.8 |
||
|
His (H) |
Class I peroxidases |
1.7 |
2.2 |
2.4 |
2.7 |
1.8 |
|
|
Class II peroxidases |
1.4 |
2.4 |
2.8 |
2.7 |
2.2 |
||
|
Class III peroxidases |
0.9 |
2.5 |
1.2 |
2.5 |
3.3 |
||
|
Ile (I) |
Class I peroxidases |
6.6 |
4.9 |
4.2 |
4 |
5.3 |
|
|
Class II peroxidases |
6.3 |
3.9 |
6.4 |
5.2 |
4.7 |
||
|
Class III peroxidases |
5.3 |
5.8 |
5.1 |
3.1 |
4.8 |
||
|
Leu (L) |
Class I peroxidases |
8.6 |
8.6 |
7.8 |
9.2 |
9.7 |
|
|
Class II peroxidases |
6.6 |
9.3 |
5.8 |
6.9 |
8.3 |
||
|
Class III peroxidases |
6.8 |
9.8 |
8.7 |
10 |
9.1 |
||
|
Lys (K) |
Class I peroxidases |
10.6 |
7.1 |
4.5 |
2.5 |
10.6 |
|
|
Class II peroxidases |
0.5 |
3 |
1.9 |
3.3 |
3.6 |
||
|
Class III peroxidases |
5.3 |
9.2 |
3.3 |
4.4 |
7.3 |
||
|
Met(M) |
Class I peroxidases |
3.4 |
1.9 |
2.6 |
1.7 |
1.5 |
|
|
Class II peroxidases |
1.9 |
3 |
0.3 |
0.8 |
0.6 |
||
|
Class III peroxidases |
4.6 |
1.2 |
1.8 |
1.3 |
2.1 |
||
|
Phe (F) |
Class I peroxidases |
5.1 |
3.4 |
5.2 |
5.5 |
4.4 |
|
|
Class II peroxidases |
6.6 |
4.5 |
7.2 |
7.1 |
6.4 |
||
|
Class III peroxidases |
6.8 |
4.9 |
6 |
4.7 |
4.8 |
||
|
Pro (P) |
Class I peroxidases |
5.7 |
5.8 |
6.6 |
7 |
4.4 |
|
|
Class II peroxidases |
8.8 |
7.2 |
8.1 |
7.7 |
7.8 |
||
|
Class III peroxidases |
3.1 |
4.6 |
3.3 |
4.7 |
5.4 |
||
|
Ser (S) |
Class I peroxidases |
6.9 |
4.7 |
3.5 |
9.7 |
5.9 |
|
|
Class II peroxidases |
6.3 |
8.7 |
9.9 |
5.3 |
5.8 |
||
|
Class III peroxidases |
7.1 |
6.7 |
9.3 |
10 |
5.1 |
||
|
Thr (T) |
Class I peroxidases |
4.9 |
4.7 |
7.5 |
5.5 |
6.2 |
|
|
Class II peroxidases |
6.3 |
3.6 |
6.6 |
7 |
6.9 |
||
|
Class III peroxidases |
5 |
5.5 |
8.1 |
7.2 |
3.9 |
||
|
Trp (W) |
Class I peroxidases |
0.9 |
1.5 |
0.5 |
1 |
0.9 |
|
|
Class II peroxidases |
0.5 |
0.6 |
0.8 |
0.3 |
0.3 |
||
|
Class III peroxidases |
0.3 |
0.3 |
0.6 |
0.6 |
0.3 |
||
|
Tyr (Y) |
Class I peroxidases |
2.9 |
4.3 |
2.6 |
3.5 |
2.1 |
|
|
Class II peroxidases |
0 |
1.2 |
0.8 |
0 |
0 |
||
|
Class III peroxidases |
1.2 |
0 |
0.8 |
0 |
0 |
||
|
Val (V) |
Class I peroxidases |
6.6 |
6 |
4 |
4.2 |
6.5 |
|
|
Class II peroxidases |
7.4 |
6 |
7.4 |
7.2 |
7.8 |
||
|
Class III peroxidases |
6.8 |
8.9 |
8.7 |
9.1 |
9.1 |
Table 3: Comparative analysis of amino acid residues in different classes of plant peroxidase.
Class I peroxidase: 1. Helicobacter pylori B8 2. Escherichia coli str. K-12 3. Rhodopirellula baltica SH1 4. Alcanivorax borkumensis SK2 5. Campylobacter coli 15-537360
Class II peroxidase: 1. Ceriporiopsis subvermispora B 2. Fomitopsis pinicola FP58527SS1 3. Ganoder malucidum 4. Schizophyllum sp. F17 5. Cerrena unicolor
Class III peroxidase: 1. Gossypium hirsutum 2. Beta vulgaris 3. Solanum lycopersicum 4. Stylosanthes humilis 5. Nelumbo nucifera
Secondary structure analysis
An average of the sequence for all three classes of peroxidase superfamily were done and checked for secondary structure analysis. All three classes of peroxidase family were rich in random coils which are known to play an important function in determining proteins flexibility and the conformational changes as turnover. Also for a protein to have higher amino acid residues in random coils is an indication for its enzymatic nature. This was in accordance with our study where all the sequence showed high percentage of random coils with class II having highest random coils and class III possessing the least [36]. In addition, all three classes showed higher number of amino acid residues in α-helix as compared to the β- turn. These secondary structures dominated by α-helix region, depicts about the thermals resistance of a protein on basis of their intrinsic stability (Figure 1) [37]. In this case class III peroxidase showed highest % of α-helix while least was depicted by class I peroxidase (Figure 1).
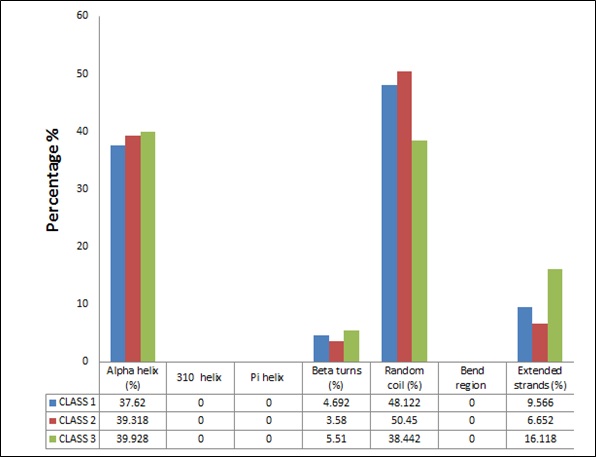
Figure 1: Comparison between secondary structures of different classes of plant peroxidase superfamily.
Tertiary structures analysis
3-D model was prepared to predict the tertiary structure for all three classes of the peroxidase superfamily. For tertiary analysis one structure was developed from each of the class using the PYRE 2 software (Figure 2). The validation of the structure was done using the Ramachandran plot. Amino-acids present in favorable and the allowed region of the Ramachandran-plot were used for the validation of the model [36].
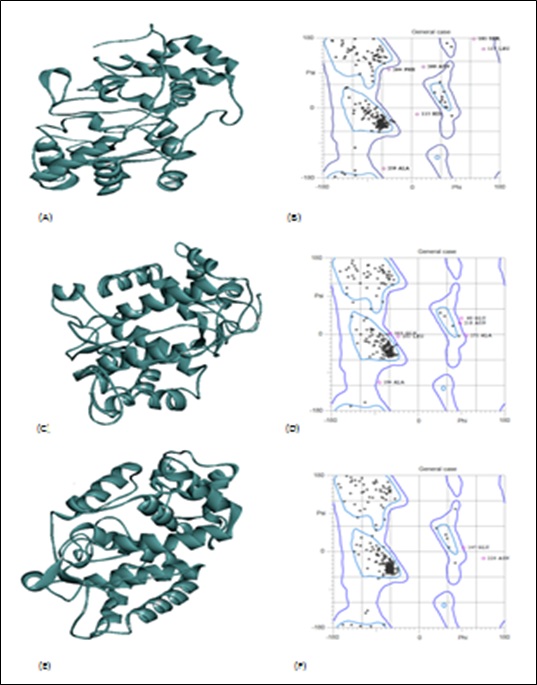
Figure 2: Class I peroxidase (CBI65628.1) (A) 3D model (B) Ramachandran Plot. Class II peroxidase (EPT05268.1) (C) 3D model (D) Ramachandran Plot. Class III peroxidase (ACJ11766.1) (E) 3D model (F) Ramachandran Plot.
Phylogenetic Analysis of Three Classes of Plant Peroxidase Superfamily
Phylogenetic tree analysis corresponds to the evolutionary distances which may help to estimate the divergence times in genes, proteins, species and in populations. The phylogeny observed showed the significant evolutionary distances for all the three classes of peroxidases (Figure 3). The class II and class III peroxidases were found to be diverged earlier from the ancestor with a shorter branch length as compared to class I. The co-ancestor for class II and class III was found to be same from the root. However, the class I peroxidases were found to be diverged from the root ancestor [37]. The phylogenetic analysis (Figure 3) also explains as to why some properties of class I peroxidase are different from rest classes under comparison as its genetically diverged before the other two classes. A classic example can be as class I peroxidase is intra-cellular in nature while class II and class III are extra-cellular in nature.
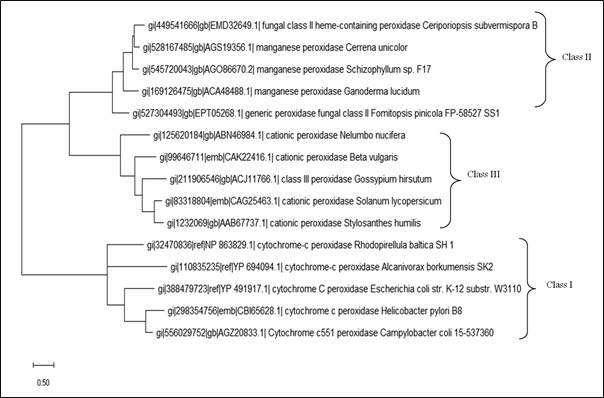
Figure 3: Phylogenetic tree of peroxidases classes obtained by MEGA-X tool. The maximum likelihood method was applied with 1000 bootstrap replications and the Jones–Taylor–Thornton model of amino acid substitutions.
In the present study, an attempt was also made to compare three classes of plant peroxidases based on their physiochemical properties, phylogeny and the predominance of specific amino acids in the individual group. This study gave vital information about characteristics in which the classes of plant peroxidases are alike and in which aspect they differ from each other [38]. The important physiochemical properties such as number of amino acids, molecular weight and aliphatic index tells us that all the three classes are somewhat similar in composition, which makes the three properties to be non-significant as overall difference does not vary. On the other hand negatively charged residues (Arg + Lys) and positively charged residues (Asp + Glu) which have long been known to function as determinants of membrane topology [38] are found to be statistically significant.
Due to diversity of 20 amino acids and due to the incredible number of combinations they afford, proteins differ widely in physiochemical properties, substrate specificity [39] as well as mechanism of their catalytic action. The plant peroxidases share similar overall protein folds and specific features, such as catalytically essential histidine and arginine residues in their active sites [9]. The non-significant value of histidine and arginine in the study showed their common presence in all three classes. Significant difference for amino acid combination were found between the classes of plant peroxidase super family, which is the basic criteria of subdivision of plant peroxidase superfamily [40]. Glycine (G) and Valine (V) amino acids present in large quantity in class III are responsible for compact core packing and functional regulation [41,42]. Cystine content was exponentially high in class II peroxidases. Cystine is said to be responsible for imparting stability to the class II peroxidase as cysteine and methionine forms disulphide bonds which affect the stability of the proteins [43]. Cysteine (C) tends to provide flexibility and is capable of making cavities in the core of the psychrophilic protein structure [44] which imparts extra stability to proteins. Asparagine (Asn) and Glutamine (Gln) have interesting hydrogen-bonding properties since they resembled the backbone peptides and hence their more value for class II peroxidase showed that they were structurally more stable [45,46].The class I peroxidases contains various evolutionary unique features such as no cystine bridges, no carbohydrate, no structural Ca2+, and no endoplasmic reticulum signal sequence, whereas the class II and class III peroxidases, are routed via the endoplasmic reticulum [47,48]. The present study provided a greater insight into the functioning, similarity and dissimilarity among each other.
CONCLUSION
The members of the plant peroxidases superfamily were compared and contrasted on the basis of their amino acid sequences and physiochemical properties using bioinformatics tools. After comparison, it was observed that three major classes of plant peroxidase superfamily mainly differ in many physiological properties such as theoretical pI, presence of positive and negative charged amino acid and composition of amino acids constituting the protein. This study has clearly worked out prominent differences between three major classes of peroxidases in conserved amino acid residues at several points.
The significant differences in the total number of amino acid residues among these classes were sufficient to determine factors like stability and nature (charge) of the enzyme. It was observed that class III was most stable among plant peroxidase superfamily, and class I being the least stable. The study helped to get greater knowledge about how closely these classes are related to each other structurally. The phylogenetic studies has signified that class II and class III peroxidase are evolutionary more closely related to each other as compared to class I peroxidase. The pI index being higher for the class I with respect to the other two classes showed that negative amino acids are present in abundance in class I peroxidases when compared with other two classes. Distribution of amino acids also helped in inferring the intracellular nature of class I plant peroxidases, which differed from rest two classes of the plant peroxidase families being extracellular in nature. The results of the present study will indeed be of great help to get an insight of the peroxidase plant superfamily, to understand the role of amino acids to develop practical strategies in engineering these peroxidases and their potential use in different industries, their role in biological and in bioremediation processes.
ACKNOWLEDGMENTS
This work has been funded by Council for Scientific and Industrial Research, New Delhi, under a CSIR-NET Junior Research Fellowship [File No.09/237(0161)/2017-EMR-1] awarded to one of the authors (VC). The authors are thankful to CSIR, New Delhi as well as DBT, New Delhi for continuous financial support to the Bioinformatics Centre, Himachal Pradesh University, Shimla (India).
CONFLICT OF INTEREST
Authors declare that they have no conflict of interest
REFERENCES
- Pandey VP, Awasthi M, Singh S, Tiwari S, Dwivedi UN (2017) A comprehensive review on function and application of plant peroxidases. Biochem Anal Biochem ISSN: 2161-1009.
- Manu BT and Rao UJSP (2009) Calcium modulated activity enhancement and thermal stability study of a cationic peroxidase purified from wheat bran. Food Chem 114: 66-71.
- Tayefi-Nasrabadi H, Dehghan H, Daeihassani B, Movafegi A, Samadi A (2011) Some biochemical properties of guaiacol peroxidases as modified by salt stress in leaves of salt tolerant and salt-sensitive safflower (Carthamus tinctorius cv.) cultivars. Biotechnol J 10: 751-763.
- Falade AO, Mabinya LV, Okoh A, Nwodo UU (2019) Studies on peroxidase production and detection of Sporotrichum thermophile-like catalase-peroxidase gene in a Bacillusspecies isolated from Hogsback forest reserve, South Africa. Heliyon 5: 03012.
- Flohe L, Ursini F (2008) Peroxidase: a term of many meanings. Antioxid & Redox Signal. 10: 1485-1490.
- Pineyro MD, Parodi-Talice A, Arcari T, Robello C (2008) Peroxiredoxins from Trypanosoma cruzi: virulence factors and drug targets for treatment of Chagas disease. Gene 408: 45-50.
- Welinder KG (1985) Plant peroxidases. Eur J Biochem 151: 497-504.
- Tuncer M, Kuru A, Sahin N, Isikil M, Isik K (2009) Production and partial characterization of extracellular peroxidase produced by Streptomyces F6616 isolated in Turkey. Ann Rev Microbiol 59: 323-334.
- Welinder KG (1992) Superfamily of plant, fungal and bacterial peroxidase. Curr Opin Struct Biol 2: 388-393.
- Passardi F, Zamocky M, Favet J, Jakopitsch C, Penel C, et al (2007) Phylogenetic distribution of catalase-peroxidases: are these patches of order in chaos?. Gene 397: 101-113.
- Lakshmi S, Shashidhara GM, Madhu GM, Muthappa R, Vivek HK, et al (2018) Characterization of peroxidase enzyme and detoxification of phenols using peroxidase enzyme obtained from Zea mayswaste. App Water Sci 8: 207.
- Furtmüller PG, Zederbauer M, Jantschko W, Helm J, Bogner M, et al (2006) Active site structure and catalytic mechanisms of human peroxidases. Archives of Biochemistry and Biophysics Volume 445: 119-213.
- Wang CY (1995) Relationship between free radical scavenging enzymes and chilling tolerancein zucchini squash. Acta Hortic 398: 205-213.
- Akbudak MA, Filiz E, Vatansever R, Kontbay K (2018) Genome-wide identification and expression profiling of Ascorbate peroxidase (APX) and Glutathione peroxidase (GPX) genes under drought stress in sorghum (Sorghum bicolor). J Plant Growth Regul 37: 925-36.
- Mantha R, Biswas N, Taylor KE, Bewtra JK, May SW (1999) Applications of oxidoreductases. Curr Opin In Biotechnol 10: 370-375
- Liu JX, Feng K, Duan AQ, Li H, Yang QQ, et al (2019) Isolation, purification and characterization of an Ascorbate peroxidase from celery and overexpression of the AgAPX1gene enhanced ascorbate content and drought tolerance in Arabidopsis. BMC Plant Biol 19:
- Agostini E, Hernández-Ruiz J, Arnao MB, Milrad SR, Tigier HA, et al (2002) A peroxidase isoenzyme secreted by turnip (Brassica napus) hairy–root culture inactivation by hydrogen peroxide and application in diagnosis kits. Biotechnol Appl Biochem 35: 1-7.
- Dawkar VV, Jadhav UU, Telke AA, Govindwar SP (2009) Peroxidase from BacillusVUS and its role in the decolorization of Textile Dyes. Biotech and Bioprocess Engineering,14: 361-368.
- Sharp KH, Mewies M, Moody PCE, Raven EL (2003) Crystal structure of the ascorbate peroxidase–ascorbate complex. Nat Str Biol 10: 303-307.
- Kangasjärvi S, Lepistö A, Hännikäinen K, Piippo M, Luomala EM, et al. (2008) Diverse roles for chloroplast stromal and thylakoid-bound ascorbate peroxidases in plant stress responses. Biochem J 412: 275-285.
- Bernroitner M, Zamocky M, Furtm€uller PG, Peschek GA, Obinger C (2009) Occurrence, phylogeny, structure, and function of catalases and peroxidases in cyanobacteria. J Exp Bot 60: 423-440.
- Yamada Y, Fujiwara T, Sato T, Igarashi N, Tanaka N (2002) The 2.0 Å crystal structure of catalase-peroxidase from Haloarculamarismortui. Nat Struct Biol 9: 691-695.
- Smulevich G, Jakopitsch C, Droghetti E, Obinger C (2006) Probing the structure and bifunctionality of catalase–peroxidase (KatG). J Inorg Biochem 100: 568-585.
- Piontek K, Smith AT, Blodig W (2001) Lignin peroxidase structure and function. Biochem Soc Trans 29:111-116.
- Pérez-Boada M, Ruiz-Dueñas FJ, Pogni R, Basosi R, Choinowski T, et al (2005) Versatile peroxidase oxidation of high redox potential aromatic compounds: site-directed mutagenesis, spectroscopic and crystallographic investigation of three long-range electron transfer pathways. J Mol Biol 354: 385-402.
- Martinez M, Ruiz-Duenas FJ, Guillen F, Martinez AT (1996) Purification and catalytic properties of two manganese peroxidase isoenzymes from Pleurotuseryngii. Eur J Biochem 237: 424-432.
- Tognolli M, Penel C, Greppin H, Simon P (2002) Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129-138.
- Cosio C, Dunand C (2009) Specific functions of individual class III peroxidase genes. J Exp Bot 60: 391-408.
- Passardi F, Penel C, Dunand C (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534-540.
- Barceló AR, Pomer F (2001) Oxidation of cinnamyl alcohols and aldehydes by a basic peroxidase from lignifying Zinnia elegans hypocotyls. Phytochem 57: 1105-1113.
- Bernards MA, Fleming WD, Llewellyn DB, Priefer R, Yang X, et al. (1999) Biochemical characterization of the suberization-associated anionic peroxidase of potato. Plant Physiol 121:135-146.
- Gazaryan IG, Lagrimini LM, Ashby GA, Thorneley RN (1996) Mechanism of indole-3-acetic acid oxidation by plant peroxidases: anaerobic stopped-flow spectrophotometric studies on horseradish and tobacco peroxidases. Biochem J 313: 841-847.
- Allison SD, Schultz J (2004) Differential activity of peroxidase isozymes in response to wounding, gypsy moth and plant hormones in northern red oak. J Chem Ecol 30: 1363-1379.
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR., et al (2005) Protein Identification and Analysis Tools on the ExPASy Server;(In)John M. Walker (ed): The Proteomics Protocols Handbook, Humana Press 571-607
- Geourjon Cand, Deléage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11: 681-684.
- Dutta B, Banerjee A, Chakraborty P, Bandopadhyay R (2018) In silico studies on bacterial xylanase enzyme: Structural and functional insight. Functional interaction of genes under Peroxidase superfamily need to be studied. J Gen Eng Biotec 16: 749-756.
- Sarkara S, Banerjee A, Chakraborty N, Soren K, Chakraborty P, et al (2019) Structural-functional analyses of textile dye degrading azoreductase, laccase and peroxidase: A comparative in silico Elect J Biotech 43: 48-54.
- Liszkay A, Kenk B, Schopfer P (2003) Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth. Planta 217: 658-667.
- Huang R, Xia R, Hu L, Lu Y, Wang M (2007) Antioxidant activity and oxygen-scavenging system in orange pulp during fruit ripening and maturation. Sci Hortic 113: 166-172.
- Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, et al (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47: 851-863.
- Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, et al (1993) The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14:1023-1031.
- Andersson H, Bakker E, and Heijne GV (1992) Different positively charged amino acids have similar effects on the topology of a polytopic transmembrane protein in Escherichia coli. The Journal of Biological Chemistry 267: 1491-1495.
- O’Reilly C, Turner PD (2003) The nitrilase family of CN hydrolyzing enzymes – a comparative study. J Appl Microbiol 95: 1161-1174.
- Yeom SJ, Kim HJ, Lee JK, Kim DE, Oh DK (2008) An amino acid at position 142 in nitrilase from Rhodococcusrhodochrous ATCC 33278 determines the substrate specificity for aliphatic and aromatic nitrile. Biochem J 415: 401-407.
- Welinder KG (1992) Superfamily of plant, fungal and bacterial peroxidases. Curr Opin Struct Biol 2: 388-393.
- Kumwenda B, Litthauer D, Bishop ÖT, Reva O (2013) Analysis of protein thermostability enhancing factors in industrially important Thermusbacteria species. Evol Bioinfor 9: 327-342.
- Panja AS, Bandopadhyay B, Maiti S (2015) Protein thermostability is owing to their preferences to non-polar smaller volume amino acids, variations in residual physico-chemical properties and more salt-bridges. PLoS One 10: 0131495.
- Kim G, Weiss SJ, Levine RL (2014) Methionine-Oxidation and Reduction in Proteins. Biochim Biophys Acta 1840: 901-905.
Citation: Chauhan V, Kumari V, Kanwar SS (2020), Comparative analysis of amino acid sequence diversity and physiochemical properties of peroxidase superfamily. J Protein Res Bioinform 2: 003.
Copyright: © 2020 Vandna Kumari, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.