
Isolation of Salmonella and E. coli (E. coli O157:H7) and its Antimicrobial Resistance Pattern from Bulk Tank Raw Milk in Sebeta Town, Ethiopia
*Corresponding Author(s):
Matios LakewDepartment Of Veterinary Microbiology, National Animal Health Diagnostic And Investigation Center, Sebeta, Ethiopia
Tel:+251 942076332,
Email:matioslakew@gmail.com
Abstract
The present study assessed the occurrence and antimicrobial susceptibility patterns of Escherichia coli, E. coli O157:H7 and Salmonella species from raw milk collected from dairy cattle farms and collector’s bulk tank in Sebeta town, Ethiopia. A total of 142 milk samples were collected for bacterial isolation and identification by using conventional bacteriological techniques and BIOLOG identification system. The identified E. coli isolates were tested for antimicrobial susceptibility pattern using four different types of antibiotics by disc diffusion method. The prevalence of E. coli was 14/142 (9.9%, 95% CI = 4.9%-14.8%), all the samples were negative for E. coli O157:H7 and Salmonella enteric was isolated from one milk sample 1/142 (0.7%). The frequency of E. coli isolation was higher in those milk samples collected from milk that was stored and transported in plastic containers (12.16%) than samples from those containers made of stainless steel (7.35%), however the difference was not statistically significant ( P>0.05). While comparing the prevalence of E. coli between samples collected directly from individual dairy farms and milk collectors, statistically significant difference (P=0.028) was observed, higher E. coli prevalence was recorded in samples obtained from milk collectors (15.3%) as compared with samples collected from individual dairy farms bulk tank milk (4.3%). All E. coli isolates were found to be 100% susceptible to gentamicin followed by amoxicillin (92.9%), sulphamethoxazole-trimethoprim (92.9%) and tetracycline (85.7%). Two of the isolates showed multiple drug resistance to two drugs. These findings showed that raw milk from dairy cattle farms and collector’s bulk tank in Sebeta town was contaminated with public health important bacterial species like E. coli and Salmonella species and the observed resistant to certain antimicrobial drugs also needs attention. To ensure the quality of raw milk, stakeholders engaged in milk and dairy production chain should be trained on hygienic practices.
Keywords
Antimicrobial susceptibility; Escherichia coli; Escherichia coli O157:H7; Milk; Salmonella enteric; Sebeta
INTRODUCTION
Milk and milk products have important role in feeding the rural and urban population owing to its high nutritional value. It is the most perfect single balanced food of high biological value in nature as it contains almost all ingredients of food in right proportion and in any easily digestible form [1].
Milk is virtually a sterile fluid when secreted into alveoli of udder. Microbial quality of milk refers to the cleanness of milk. This is defined by a number of bacteria present in milk. The high bacteria count as well as the presence of pathogenic bacteria in milk not only degrades the milk quality and shelf-life of milk or milk related products but also poses a serious health threat to consumers [2]. Bacteria, yeasts and moulds are the common contaminants of milk. Their rapid growth of microorganisms, particularly at high ambient temperature can cause marked deterioration in quality of the milk and dairy products manufactured from it [3]. Microbial contamination might generally occur from three main sources: within the udder, exterior to the udder and from the surface of milk handling and storage equipment, but the surrounding air, feed, soil, faeces and grass are also possible sources of contamination [4].
The demand of consumers for safe and high quality milk has placed a significant responsibility on dairy producers, retailers and manufacturers to produce and market safe milk and milk products [5]. The safety of dairy products with respect to food-borne diseases is of great concern around the world. Raw milk can harbor dangerous microorganisms which may pose serious health risks to humans. Over 200 known diseases are transmitted through eating food contaminated by a variety of agents including bacteria, parasite, viruses, and fungi [6]. Some of the bacteria involved in causing food borne diseases due to the consumption of raw milk include Escherichia coli, Listeria monocytogenes, Salmonella, Campylobacter, Brucella abortus, Staphylococcus aureus, Bacillus cereus, Mycobacterium spp. and Clostridium botulinum. If these pathogenic bacteria are present in raw milk, it is a major public health concern, especially for those individuals who drink raw milk frequently [7].
Over the last 20 years, the emergence of major food borne pathogens such as Salmonella and Escherichia coli have persisted as a major public health concerns and provide clear examples of the persistence of food borne pathogens despite considerable efforts aimed at prevention and control [8]. For this reason, the basic steps in the control of safety and quality of food include analysis of food products for presence of pathogenic microorganisms that cause the majority of alimentary human diseases. Among them, Salmonella and E. coli O157:H7 are the major ones. These food borne pathogens have frequently been linked to a number of cases of human illness [9].
The increasing use of antibiotics in veterinary practice is suspected to contribute to acceleration of antibiotic resistance in microorganisms [10]. The irrational use of antibiotics in food producing animals could result into antibiotic residues in edible tissues and products [11]. It has been reported that, antibiotics used for treatment of human bacterial infections are used for prophylactic, therapeutic and growth promotion in animals too [12]. Bacteria that have been exposed to low doses of these antibiotics in tissues and products from these animals may be less susceptible to drugs, and when such bacteria enter the human body through consumption of contaminated foods, they may cause infections that are resistant to many antibiotics [13].
Gastroenteritis due to food-borne disease is one of the most common illnesses in Ethiopia, and it is a leading cause of death among people of all ages in the country [14]. The lack of surveillance of food-borne pathogens, poor hygienic conditions and the wide spread cultural practice of raw milk consumption are all major factors contributing to the high risk of exposure of Ethiopians to food-borne pathogens such as E. coli, E. coli O157:H7 and Salmonella species. In spite of the high risk of exposure to E. coli, E. coli O157:H7 and Salmonella from animal origin food sources like milk in the country, there is lack of well-organized report indicating the contamination level of animal origin food sources like milk by these zoonotic organisms. Moreover, in the country both veterinary and medical drugs are often misused, creating ideal conditions for the development of resistant strains, thus better understanding of the antimicrobial susceptibility/resistance/ patterns of pathogens isolated from animal source foods like milk is needed. Therefore the objectives of this study were:-
- • To isolate coli with emphasis on E. coli O157:H7 and Salmonella species from raw milk obtained from dairy cattle farms and milk collectors in Sebeta town, Ethiopia.
- • To determine antimicrobial susceptibility and resistance pattern of E. coli isolates from milk samples.
MATERIALS AND METHODS
Study AREA
The study was conducted in Sebeta town South West Showa, from November 2017 to March 2018. The mean annual temperature and rainfall ranges between 15°C to 21°C and 800 mm to 1199 mm respectively and it is located 25 km west of Addis Ababa. Intensive and semi-intensive cattle dairy farms with exotic and cross breeds are managed by the community at the area
Study design
A cross-sectional study was conducted to determine the prevalence of E. coli with emphasis on E. coli O157:H7 and Salmonella species. Milk samples were collected from individual dairy cattle farms and milk collectors from Sebeta town.
Sample size determination
The approximate sample size required for the study was determined based on the expected prevalence of E. coli and the desired absolute precision using the formula stated on Thrusfield [15].
n= [1.962 Pexp (1- Pexp )] / d2
Where: n=required sample size ; Pexp =Expected prevalence ; d = desired absolute precision
The previous study made in Holeta and Burayu by Yohannes [16] showed the prevalence of E. coli was 7.1% in cow milk. Therefore, by using this 7.1% expected prevalence, at a confidence level of 95% and required absolute precision of 5%, the calculated minimum sample size was 128 and 142 bulk tank milk samples were collected to increase the precision.
STUDY METHODOLOGY
Sampling methods and procedures
General information was collected from Sebeta town livestock and fisher office to identify the total number of farms, farm size, farming system, and the status and number of milk collectors in Sebeta town. According to the result the majority of the farms were at the household/smallholder level, with farm size not more than 5 cows per farm. Milk collecting site at the main road were identified as main sources of milk for consumers and processing center and included in the study. Simple random sampling technique was applied to collect raw milk samples from each group of collecting site and farms bulk tank. Milk samples were aseptically taken in morning time. The reason for collecting morning milk samples was, most of dairy farms submit their milk to milk collectors in the morning time soon after milking. During collection, approximately about 4-5 ml raw milk samples were aseptically collected from bulk tank milk container of collectors and dairy farms, then placed in sterile universal bottle by using sterile graduated pipette for each samples. Subsequently these samples were labeled and immediately transported to National Animal Health Diagnostic Center (NAHDIC) to isolate E. coli, E. coli O157:H7 and Salmonella species from raw cow milk.
Isolation and identification of E. coli and E. coli O157:H7
Each raw cow milk samples were inoculated on MacConkey agar, and then incubated at 370C for 24 hours. Typical colonies on MacConkey agar (pink, due to their ability to ferment lactose) were stained using gram stain and observed for their staining and morphological characteristics and transferred to Eosin Methylene- Blue (EMB) agar. The colonies with green metallic sheen on EMB agar which is typical feature of E. coli were transferred to sorbitol MacConkey agar to check the presence of E. coli O157:H7 phenotype (inability to ferment sorbitol). Then the E. coli suspected colonies were transferred to nutrient agar to be used for secondary biochemical tests (IMViC tests) [17]. A standard reference strain of E. coli (ATCC 25922) was used as a quality control.
Based on Primary and secondary biochemical tests E. coli suspected colonies were confirmed by BIOLOG bacterial identification system. Then, BIOLOG system (fully automated coated microplate based bacterial identification system) using GEN III micro plate (Lot number 3003241, BIOLOG, USA) with protocol A method was used to further confirm the species of suspected colonies. A single colony grown on Biolog Universal Growth (BUG) agar medium was selected and emulsified into ‘Inoculating Fluid A’ (IF A). According to the manufacturer's instructions, cell density of the bacterial inoculum was measured and adjusted for a specified transmittance (90 to 98%) using a turbidimeter. For each isolate, 100 μl of the bacterial cell suspension was inoculated in to each of the 96 well coated micro plates, using automatic multichannel pipette and incubated aerobically at 330C for 22 hr. BIOLOG microstation reader was used to read the incubated microplate and provides species/sub-species Identification (ID), and then the results were printed out [18].
Isolation of salmonella species
Salmonella species isolation was taken based on NAHDIC’s test method for Salmonella species identification which was adapted from ISO 6579-1:2017 [19]. Briefly, homogenized raw milk sample of 1 ml was added to 9ml of sterilized buffered peptone water and incubated overnight at 37°C. Then for selective enrichment 0.1ml of pre-enrichment was transferred to 10 ml of Rappaport Vassiliadis Soya broth (RVS broth) then were incubated at 41.5 ºC for 24 hrs. Each selective enrichment broth bottle was well shaken and then a loop full from each was streaked onto plates of Xylose Lysine Deoxycholate (XLD) agar and all plates were then aerobically incubated at 37°C for 24 hrs. Pink colonies with or without black centers were transferred to nutrient agar for further test. Then secondary biochemical tests (IMViC tests), TSI, urea and lysine were conducted. A standard reference strain of Salmonella Typhimurium ATCC14028 was used as a quality control. Finally Salmonella species suspected colonies were confirmed by using GEN III microplate, BIOLOG system.
Antimicrobial susceptibility testing for E. coli
The antimicrobial susceptibility test was performed following the standard agar disk diffusion method using commercial antimicrobial disks. The selection criteria of the antibiotics depended on the regular use of the antimicrobials in the animal and human treatments. Mueller-Hinton agar media was used for susceptibility testing. The isolated E. coli strains were tested for sensitivity to commonly used antimicrobials in veterinary medicine in the country including Gentamicin (GCN) (10????g), Trimethoprim-sulfamethoxazole (SXT) (25????g), Tetracycline (TE) (30????g) and Amoxicillin 25µg. A standard reference strain of E. coli (ATCC 25922) was used as a quality control. Interpretation of results was made according to CLSI Guideline [20].
DATA MANAGEMENT AND ANALYSIS
Microsoft excel spread sheet was employed for raw data entry and SPSS version 20.0 software was used for descriptive statistics. For all analysis, 95% CI and P-value<0.05 was set for statistical significance of an estimate.
RESULTS
Prevalence of E. coli, E. coli O157:H7 and Salmonella species from bovine raw bulk tank milk
A total of 142 milk samples were collected from dairy cattle farms and milk collectors’ bulk tank for isolation and identification of E. coli, E. coli O157:H7 and Salmonella species. Prevalence of E. coli and Salmonella species are summarized in table 1. E. coli and Salmonella species were detected in 14/142 (9.9%) and 1/142 (0.7%) raw milk samples, respectively. Based on the other finding E. coli O157:H7 was not isolated from bulk tank milk samples collected from the area.
|
Species of Bacteria |
Positive milk samples (%) |
95% Confidence Interval |
|
E. coli |
14/142 (9.9%) |
4.9-14.8% |
|
Salmonella enterica |
1/142 (0.7%) |
.0-2.1% |
Table 1: Prevalence of E. coli and Salmonella species.
E. coli prevalence in raw milk value chain was evaluated and a higher rate of contamination was detected in the samples collected from collectors (15.3%) than from dairy farms (4.3%) the difference was statistically significant (P<0.05%) (Table2). The container in which milk was collected was also evaluated and a higher frequency of contamination was detected in the milk samples collected from plastic containers (12.16%) than stainless steel (7.35%). The containers made of plastic were identified to be more prone to be contaminated by E. coli than stainless steel, but the difference was not statistically significant (P>0.05%).
|
Source of milk sample |
Number of samples tested |
Positive samples |
Percentage of E. coli isolation from milk samples |
Chi square |
P-value |
|
From farm |
70 |
3 |
4.3 |
4.825 |
0.028 |
|
From collectors |
72 |
11 |
15.3 |
|
|
|
Type of container |
|||||
|
Plastic container |
74 |
9 |
12.16 |
0.922 |
0.337 |
|
Metal/Stainless steel/ milk can |
68 |
5 |
7.35 |
|
|
Table 2: E. coli prevalence in raw milk value chain. Antimicrobial susceptibility/resistance pattern of E. coli isolated from bovine bulk tank milk
A total of 14 E. coli isolates were tested against using 4 antimicrobial discs following CLSI guidelines. All E. coli isolates were found to be 100% susceptible to gentamicin followed by amoxicillin (92%) and sulphamethoxazole-trimthoprim (92%) and then tetracycline (85%), figure 1. One isolate showed resistance for amoxicillin and another for sulphamethoxazole-trimethoprim and two isolates were resistant for tetracycline. Two of the isolated E. coli species showed multiple resistances to two drugs (one isolate for Amoxicillin-Tetracycline and the other for Tetracycline-Sulpha-Trimethoprim).
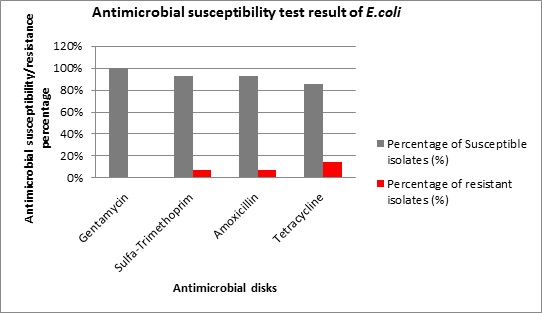
Figure 1: Antimicrobial susceptibility test result of E. coli.
DISCUSSION
Milk, a perishable complete nutritious food is considered to be a good medium of growth for many of the microorganisms [21]. E. coli is a normal inhabitant of the intestines of animals and humans. However, its recovery from food may be of public health concern due to the possible presence of enter- pathogenic and/or toxigenic strains like E. coli O157:H7 which can lead to sever gastro intestinal disturbances [22] and other life threatening syndromes on the consumer [23].
Escherichia coli and salmonella are not only regarded as an indicator of fecal contamination but more likely as an indicator of poor hygiene and sanitary practices during milking and further handling. In this study, a total of 142 raw milk samples were studied and from these 14(9.9%) milk samples were contaminated with E. coli, other finding E. coli O157:H7 was not isolated (0%) and Salmonella enterica was identified from 1(0.7%) sample. The isolated percentage of E. coli is in agreement with the report by [16,24] who reported 11.6% and 7.1% respectively. On the other hand, the present study was relatively lower as compared to the studies by [25-28] previously reported as 26.57%, 27.91%, 25% and 33.9% respectively. The other finding of the present study is that Escherichia coli O157:H7 was not isolated (0%) from bulk tank milk samples. Many other studies on the prevalence of E. coli O157:H7 from raw milk reported that the isolation rate was low and varied between 0 % and 10% [29]. It has been suggested that the microorganisms are not actually excreted in the milk but probably result from faecal contamination of milk during the milking process [30].
Salmonella species cause enteric infection characterized mainly by gastroenteritis on humans and other animals worldwide, and sometimes in severe cases it can result in systemic infection and even death. In general, Salmonella prevalence observed in study was 1/142 (0.7%). Other studies [31-33] reported prevalence of 0%. Moreover study at Jigjiga City of Somali Regional State also reported 1 (3.3%), lower percentage of Salmonella species [34]. Moreover, several studies recorded that Salmonella was not detected in milk samples [35-38]. On the other side relatively higher percentages of 8.7% in Nigeria [39] and 20% in Ethiopia was reported [40].
The variation that was seen in prevalence of E. coli and Salmonella species in different studies may be due to difference in sample size, farming system, farm size, milking equipment, milking technique, geography, ecology, duration of milk transportation, and hygienic conditions [22]. The presence of E. coli may not necessarily indicate a direct fecal contamination of milk but is an indicator of poor hygiene and unsanitary practices during milking and further handling of milk and presents a potential hazard for people consuming such products [41]. Moreover, bacterial identification techniques used by different researches may also be one factor where using only primary and secondary biochemical tests by most of the authors and other limited studies further confirm the isolates by techniques like BIOLOG, this reduces the number of positive samples.
Raw milk in value chain is commonly distributed locally to consumers with no controlled measures to maintain the safety and quality before it reaches consumers in Sebeta. The prevalence of E. coli was different at the raw milk chain. Difference in prevalence of E. coli was observed on the source of raw milk samples, from farm and collectors. Higher prevalence was recorded in collector’s raw bulk tank milk (15.3%) as compared to milk samples from individual dairy farms (4.3%) raw bulk tank milk. The observed high prevalence of E. coli from milk collectors was higher than those previous reports from Holeta and Sululta farmers 3.84% and 11.53% [31]. The observed differences might be due to the longer time for transporting milk to the collectors at ambient temperature under poor hygienic conditions which support the growth of the bacteria in the milk samples taken from the collectors. Moreover the collectors receive milk from several individual dairy farms and there is no well organized checking system for quality of milk during collection.
The container in which milk was collected was also evaluated and a higher rate of contamination was detected in the samples collected from milk held in plastic containers (12.16%) than stainless steel (7.35%). The containers made of plastic were identified to be more prone to be contaminated by E. coli than stainless steel, but the difference was not statistically significant (p>0.05). Similar findings were also reported before in the country [28].
The development of antimicrobial resistance among the pathogenic bacteria poses a problem of high concern. The present study showed that E. coli isolates were highly sensitive to gentamicin followed by amoxicillin and sulphamethoxazole-trimthoprim (92%) and then tetracycline (85%). Similarly other studies from Mekele town [26] revealed that the susceptible to some antibiotics like gentamicin (100%) and tetracycline (60%). Moreover, other study [27] has also reported all the isolates were found to be 100% susceptible to gentamicin and sulphamethoxazole-trimthoprim (76%). Multiple drug resistance patterns were also analysed, accordingly, 2(14.28%) of the E. coli isolates have shown resistance to two of the antimicrobials tested. In particular, these bacterial isolates have primarily shown multidrug resistance to Amoxicillin-Tetracycline and the other for Tetracycline-Sulpha-Trimethoprim. Previous studies have also reported the development of multiple drug resistance by E. coli [42,43]. In general, the development of antimicrobial resistance could possibly be due to overuse and misuse of the antimicrobials by herd owners and animal health professionals. Furthermore, even the undergone bacterial mutations may be the cause for the development of antimicrobial resistance. Drugs like tetracycline are commonly used in the country for treatment and disease prevention in animal health sector of the country. Some studies describe the direct correlation of antimicrobial use to antimicrobial resistance in veterinary medicine at a supranational level, based on publicly available data sources [44]. Thus, the observed proportion of resistance of E. coli isolates to tetracycline may be related with the widespread usage of the drug in the country.
CONCLUSION
The findings obtained in this study revealed that raw cow’s milk were found to be contaminated with E. coli and salmonella species. The sources of E. coli and salmonella in the raw cow milk may be from contaminated udders, contaminated water, poor sanitation practices, contaminated containers, and milk handlers themselves. Since the milk is transported and managed at an ambient temperature, high microbial populations can be reached within short period of time. Based on the antimicrobial susceptibility pattern, most of the E. coli isolates were found to be susceptible to gentamicin, amoxicillin, sulphamethoxazole-trimethoprim and tetracycline and few isolates have also showed resistance. Accordingly, to ensure the quality of raw milk, every actor engaged in milk and dairy production chain should be trained for hygienic practices. Proper antibiotics usage in animal production also needs attention.
COMPETING INTERESTS
The authors declare that they have no competing interests.
ACKNOWLEDGEMENT
The authors acknowledge National Animal Health Diagnostic and Investigative Center (NAHDIC), mainly for supporting all the laboratory consumables used for sample processing and staffs of General bacteriology laboratory were helpful during bacterial isolation, identification and antimicrobial susceptibility test. Moreover, the authors appreciate University of Gondar College of Veterinary Medicine and Animal Science (UoG/CVMAS) for advisory and technical support while conducting the research. In addition, the authors appreciate the cooperation of the dairy farm owners and milk collectors at Sebeta town during sample collection.
REFERENCES
- Khaliq K, Ashfaque M, Hussain I, Akhtar M (2001) Bacteriological studies on raw milk supplied to Faisalabad city during summer months. University of agriculture, Faisalabad, Pakistan.
- Yuen SK, Yee CF, Yin FH (2012) Microbiological Quality and the Impact of Hygienic Practices on the Raw Milk Obtained from the Small-scale Dairy Farmers in Sabah, Malaysia. International Journal of Agricultural and Food Sciece 2: 55-59.
- FAO (1989) Animal production and health paper. Food and Agriculture Organization of the United Nations, Rome, Italy.
- Mosu S, Megersa M, Muhie Y, Gebremedin D, Keskes S (2013) Bacteriological quality of bovine raw milk at selected dairy farms in DebreZeit town, Ethiopia. Journal of Food Sciences and Technology Research 1: 1-8.
- Mennane Z, Ouhssine M, Khedid K, Elyachioui M (2007) Hygienic quality of raw cow’s milk feeding from domestic Waste in Two Regions in Morocco. International Journal of Agricultural Biology 9: 1560-8530.
- Oliver SP, Jayarao BM, Almeida RA (2005) Foodborne Pathogens in Milk and the Dairy Farm Environment: Food Safety and Public Health Implications. Foodborne Pathog Dis 2: 115-129.
- Chye FY, Abdullah, Ayob MK (2004) Bacteriological Quality and Safety of Raw Milk in Malaysia. Food Microbiology 21: 535-541.
- Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, et al. (2010) Food-borne diseases - the challenges of 20 years ago still persist while new ones continue to emerge. Int J Food Microbiol 1: 3-15.
- Brown MH, Gill CO, Hollingsworth J, Nickelson R II, Seward S, et al. (2000) The role of microbiological testing in systems for assuring the safety of beef. Int J Food Microbiol. 62: 7-16.
- Addis Z, Kebede N, Worku Z, Gezahegn H, Yirsaw A, et al. (2011) Prevalence and antimicrobial resistance of Salmonella isolated from lactating cows and in contact humans in dairy farms of Addis Ababa: a cross sectional study. BMC infect Dis 11: 222.
- Darwish, S, Eldaly A, El-Abbasy MT, Ikenaka Y, Nakayama S, et al. (2013) Antibiotic residues in food: the African scenario. Jpn J Vet Res 61: 13-22.
- Phillips I, Casewell M, Cox T, De groot B, Friis C, et al. (2004) Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J Antimicrob Chemother 53: 28-52.
- Clauben M, Bahmann D, Schmidt SD (2013) Detection of antibiotic residues in food–pitfalls and optimization of agar diffusion tests in comparison with commercial test kits. Allen Institute for AI, Washington, USA.
- Institute for Health Metrics and Evaluations (2013) The Global Burden of Disease: Generating Evidence, Guiding Policy, Washington, USA.
- Thrusfield M (2005) Veterinary Epidemiology, 3rded. Blackwell Science Ltd, Osney Mead, UK.
- Yohannes E (2016) Characterization of drug resistance patterns of coli isolated from milk collected from small scale dairy farms reared in Holeta and Burayu, and meat from Addis Ababa abattoirs enterprise and alema farm slaughter slab. Addis Ababa Uiversity Libraries, Ethiopia.
- Quinn PJ, Carter ME, Markey B, Carter GR (2013) Clinical Veterinary Microbiology, Mosby, Missouri, USA.
- OMNILOG (2010) OMNILOG data collection software, bacterial and fungi identification system, user guide part no. 90311, 2:.3.
- ISO 6579-1 (2017) Microbiology of the food chain — Horizontal method for the detection, enumeration and serotyping of Salmonella — Part 1: Detection of Salmonella spp. International Standard for organization, Geneva, Swizerland.
- Clinical Laboratory Standards Institute (2010) Performance Standards for Antimicrobial Disk Susceptibility Tests (13th edn). Clinical Laboratory Standards Institute 38: 1-94.
- Khayal AA, Ragia OM (2013) Biochemical and microbiological evaluation of fermented camel milk. New York Science Journal 6: 74-79.
- Soomro AH, Arain MA, Khaskheli M, Bhutto B (2002) Isolation of Escherichia Coli from Raw Milk and Milk Products in Relation to Public Health Sold under Market Conditions at Tandojam, Pakistan. Pakistan Journal of Nutrition 1: 151-152.
- Kawano K, Okada M, Haga T, Maeda K, Goto Y (2008) Relationship between pathogenicity for humans and stx genotype in Shiga toxin-producing Escherichia coli serotype O157. Eur J Clin Microbiol Infect Dis 27: 227-232.
- Ayano AA, Hirko F, Simayalew AM, Yohannes A (2013) Prevalence of subclinical mastitis in lactating cows in selected commercial dairy farms of Holeta district. Journal of Veterinary Medicine and Animal Health 5: 67-72.
- Sori H, Zerihun A, Abdicho S (2005) Dairy cattle mastitis in and around Sebeta, Ethiopia. International Journal Applied Research Veterinary Medicine 3: 332-338.
- Tadesse HA, Gidey NB, Workelule K, Hailu H, Gidey S, et al. (2018) Antimicrobial Resistance Profile of coli Isolated from Raw Cow Milk and Fresh Fruit Juice in Mekelle, Tigray, Ethiopia. Veterinary Medicine International 2018: 1-8.
- Yohannes G (2018) Antimicrobial Susceptibility Testing of Escherichia Coli Isolated from Selected Dairy Farms in and Around Mekelle, Ethiopia. Journal of Veterinary Science & Technology 9: 1-5.
- Disassa N, Sibhat B, Mengistu S, Muktar Y, Belina D (2017) Prevalence and Antimicrobial Susceptibility Pattern of coli O157:H7 Isolated from Traditionally Marketed Raw Cow Milk in and around Asosa Town, Western Ethiopia. Veterinary Medicine International 2017: 1-8.
- Padhye NV, Doyle MP (1991) Rapid procedure for detecting enterohemorrhagic Escherichia coli O157 in food. Appl Environ Microbiol 57: 2693-2698.
- Heuvelink AE, Van den Biggelaar FL, Zwartkruis-Nahuis J, Herbes RG, Huyben R, et al. (1998) Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J Clin Microbiol 36: 3480–3487.
- Amistu K, Degefa T, Melese A (2015) Assessment of Raw Milk Microbial Quality at Different Critical Points of Oromia to Milk Retail Centers in Addis Ababa. Food Science and Quality Management 38: 2-5.
- Mhone TA, Matope G, Saidi PT (2012) Detection of Salmonella spp., Candida albicans, Aspergillusspp and antimicrobial residues in raw and processed cow milk from selected small holder farms of Zimbabwe. Veterinary Medicine International 2012: 1-6.
- Jalali M, Abedi D, Pourbakhsh SA, Ghoukasin K (2008) Prevalence of salmonella spp. in raw and cooked foods in Isfahan? Journal of Food Safety 28: 442-452.
- Reta MA, Bereda TW, Alemu AN (2016) Bacterial contaminations of raw cow’s milk consumed at Jigjiga City of Somali Regional State, Eastern Ethiopia. International Journal of Food Contamination 3: 1-9.
- Ekici K, Bozkurt H, Isleyici O (2004) Isolation of some pathogens from raw milk of different milch animals. Pakistan Journal of Nutrition 3: 161-162.
- D’Amico DJ, Groves E, Donnelly CW (2008) Low incidence of foodborne pathogens of concern in raw milk utilized for farmstead cheese production. Journal of Food Protection 71: 1580-1589.
- Zeinhom MMA, Abdel-Latef GK (2014) Public health risk of some milk borne pathogens. Beni-Suef University Journal of Basic and Applied Sciences 3: 209-215.
- Elbagory AM, Emansh E, Eman KF (2015) Impact of probiotic strains on growth of some food poisoning bacteria from milk and soft cheese. Nutrition and Food Technology: Open Access 1: 1-6.
- Karshima NS, Pam VA, Bata S, Dung PA, Paman ND (2013) Isolation of Salmonella Species from Milk and Locally Processed Milk Products traded for Human Consumption and Associated Risk Factors in Kanam, Plateau State, Nigeria. Journal of Animal Production Advances 3: 69-74.
- Tadesse T, Dabassa A (2012) Prevalence and Antimicrobial Resistance of Salmonella Isolated from Raw Milk Samples Collected from Kersa District, Jimma Zone, Southwest Ethiopia. Journal of Medical Sciences 12: 224-228.
- Meshref AMS (2013) Bacteriological quality and safety of raw cow’s milk and fresh cream. Slov Vet Res 50: 21-30.
- Orrett FA, Shurland SM (2001) Prevalence of resistance to antimicrobial of coli isolates from clinical sources at a private hospital in Trinidad. Jpn J Infect Dis 54: 64-68.
- Kurutepe S, Surucuoglue S, Sezgin C, Gazi H, Gulay M (2005) Increasing antimicrobial resistance in Escherichia coli isolates from community-acquired urinary tract infections during 1998-2003 in Manisa, Turkey. Japanese Journal of Infectious Disease 58: 159-161.
- Chantziaras I, Boyen F, Callens B, Dewulf J (2014) Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: a report on seven countries. Journal of Antimicrobial Chemotherapy 69: 827-834.
Citation: Dadi S, Lakew M, Seid M, Koran T, Olani A, et al. (2020) Isolation of Salmonella and E. coli (E. coli O157:H7) and its Antimicrobial Resistance Pattern from Bulk Tank Raw Milk in Sebeta Town, Ethiopia. J Anim Res Vet Sci 4: 021.
Copyright: © 2020 Solomon Dadi, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

