
Selection of a Rabies Virus Strain as a New Candidate for Vaccine Development in Thailand
*Corresponding Author(s):
Nitatpattana NCenter For Vaccine Development, Institute Of Molecular Biosciences, Mahidol University At Salaya Putthamonthon, Nakhon Pathom Province, Thailand
Tel:+1 (301) 332 22 37,
Email:narong.nit@mahidol.ac.th
Abstract
Rabies remains a severe viral zoonotic disease worldwide is always fatal once clinical symptoms appear in human and domestic animals. Furthermore, the Rabies virus is commonly transmitted to humans by infected domestic animals. There is no cure available and vaccination is the only way to prevent virus infection. The virus actively circulates in Thailand with several fatal cases and outbreaks in animal; mostly dogs have been recorded for the past decade. Additionally, Thailand import 2.5 Million doses of rabies vaccine yearly, the Thai National Vaccine Institute (Ministry of Public Health, MOPH) decided to initiate research and development on rabies vaccine to set up an effective vaccine to protect against actively circulating viral strains in it and the neighboring countries. The candidate vaccine aims to provide a local cost-effective production with a potential for international licensure.
The main objective of this study was to select a candidate vaccine rabies virus strain that comply with mandatory characteristics, accordingly to WHO’s regulations, including: the ability to efficiently growth in Vero cell line; having an Immunofluorescence companion test using monoclonal anti G protein for antigen detection, and to be one of the most common strain genotype circulating in Thailand. With such objectives online, selected rabies strain genotype sequences were analyzed and characterized by their identity to the rabies virus strain actively circulating in Thailand. The identified Rabies virus TH2 strain is largely endemic throughout Thailand; it was recognized as a potential candidate for the Rabies vaccine development by showing all required and suitable characteristics. Although, the Rabies virus TH2 strain appears genetically distant from the commercially available vaccine strain in Thailand, it should stimulate immunity and prevent most of the rabies risk due to the actively circulating dog rabies strain in Thailand.
Keywords
Fix virus; Local strain; Rabies virus; Seed selection
INTRODUCTION
Rabies is an acute viral infection of the nervous system that presents both encephalitic and paralytic clinical forms. Rabies is transmitted by the rabies virus (Rabies lyssavirus) through saliva of infected mammal (e.g. dog, cat, and domestic ungulate), rarely the human to human transmission occurs, while exceptionally the virus can be transmitted through organ or tissue transplant [1]. The Rabies lyssavirus, is a negative sense, single-stranded enveloped RNA virus of the Rhabdoviridae family presenting in a typical cylindrical morphology in electron-micrograph. The genetic information of 11,615 to 11,966 pair base is packaged as a ribonucleoprotein complex tightly bound to the viral nucleoprotein while the genome encodes for five proteins including: nucleoprotein (N), glycoprotein (G), phosphoprotein (P), matrix protein (M), and polymerase (L) [2]. Although rabies is a well-known ancient disease, its virus appears endemic but particularly in low and medium-income countries (LMIC) while human rabies continues to be an important public health problem particularly in Asia and Africa [3]. The combined annual cost for both continents is estimated to be US$ 583.5 million yearly and mostly due to post exposure prophylaxis [4]. As it’s generally observed in Thailand and LMIC, the canine appears as the primary vector of rabies virus [5]. A human case of Rabies was first identified in Thailand in the 1920s and then in canines. While felines and stray animal populations grew, rabies cases also increased with a record high of 370 people death in 1980 [6]. After several outbreaks that occurred in the 1980s, canine and human rabies decreased due to an efficient post-exposure treatments, education, mas, dog and cat vaccination campaign [6]. In 2018, Thailand presented one of its major rabies outbreaks spreading among 14 provinces ( Buri ram, Rayong, Songkhla, Surin, Trang, Nakhon Ratchasima, Prachuap Khiri khan, Phatthalung, Nong khai, Yasothon, Klalasin, Mukdahan, Tak, Surat Thani), with a mortality rate of 0.03 per 100,000 inhabitants and 17 fatal cases recorded [7]. In the meanwhile, 1,469 (15.8%) confirmed cases of rabies in animals (mostly dog and cattle) from 54 provinces where reported (Rabies Situation - 7 Oct 19 /2020 - Department of Livestock Development Ministry of Agriculture and Cooperatives). After this recent and unforeseen event, the Thai government decided to put an end to these surprising epidemic outbreaks, the cost of post exposure treatment and animal vaccination remain at a very high cost due to the need to import the anti-rabies vaccine from abroad. Therefore, the National Vaccine Institute (NVI, MOPH, and Thailand) asked the Center for Vaccine Development (Institute of Molecular Biosciences, Mahidol University) and the National Institute of Health (Thailand Department of Medical Science and MOPH) to investigate the potential for the development of a human vaccine using a vaccine strain complying with international regulation. The purpose of this study was to identify and select a rabies virus strain among the entire rabies virus isolated in Thailand and stored at the bio bank of the National Institute of Health (Thai Ministry of Medical Sciences, MOPH). These strains were chosen to exhibit specific and mandatory characteristics as recommended by the World Health Organization [8] and consistently having the ability to replicate in the Vero cell line. After this preliminary phase of identifying with high accuracy a targeted strain, the future development of inactivated human rabies vaccine will be completed under the direction of the MOPH and the Government Pharmaceutical Organization (GPO).
MATERIALS AND METHODS
Cell lines
Two cell lines commercially available from ATCC were selected and use respectively for virus amplification and virus replication effectiveness, including: a mouse cell line derived from mouse (Mus musculus) Neuroblastoma (MNA, ATCC CCL-131); and the Vero cell line derived from normal kidney cells of the adult African green monkey (Cercopithecus aethiops) (ATCC CCL-81).
Rabies virus
Fifty Rabies virus strains were provided by Thai National Institute of Health including thirteen strains from human and thirty-seven strains from canine’s brain. All the strains tested positive by Indirect Fluorescent Assay (IFA) using the FITC Anti-Rabies Monoclonal Globulin (Product number 800-092)
Indirect immunofluorescent antibody test (IFAT)
IFAT was performed using an anti-nucleoprotein monoclonal antibody, commercially available (FITC Anti-Rabies Monoclonal Globulin, Fujirebio US, Inc) following a classical previously described method [9].
Rabies virus Adaptation and Selection
The 50 rabies virus strains were propagated in Mouse Neuroblastoma (MNA) and passaged to time on this cell line at 37°C under 5% CO2. Propagated strains were then selected for their response by IFAT and, only 5 strains only showed a positivity ≥2+ by IFAT (undiluted cell suspension having ≥50% of fluorescent cell were considered as positive). The five selected strains were then propagated in MNA cell line for five or more passages and then used for the virus to be titrated by Reed and Muench method (fluorescence titration) and quantitative evaluated by “Nested RT-PCR” [10]. Ultimately, the five Rabies virus strains, showing a titer >104 Tissue Culture Infective Dose (TCID), were selected and propagated into Vero cell line kept at 5% CO2, 37 °C, and followed by 5 to 10 passages .
Deep sequencing
The RNA extraction was performed on an archived selected TH 13-42-07240 virus strains (i.e. TH2 clusters) sample isolated from canine brain in 2009 [11]. From a Vero cell infected monolayer, i.e. TH 13-42-07240 rabies virus strain tested positive by IFAT, viral RNA was extracted using QIAamp Viral RNA Mini Kit (QIAGEN, Germantown, MD). Total RNA was depleted of host genomic DNA and ribosomal RNA (rRNA) using NEBNext rRNA depletion kit (New England Biolabs, UK) following the manufacturer’s instructions, eluting the final product on in 30 ml molecular gram from the collection, previously de water. The depleted RNA was then purified to remove the enzyme using the RNeasy plus mini kit (QIAGEN, Germantown, MD), without DNase digestion and eluted in 30 ml of molecular grade water. cDNA was synthesized using Maxima H minus Double stranded cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA), following the manufacturer’s instructions.
Genotype sequence analysis:
Characterizing the viral genome, we used a random amplification, deep-sequencing approach (454 Sequencing) as described elsewhere, where 98% of reads matched the Rhabdovirus genome reference (Accession number EU293111) [12]. The multiple sequence alignment was based on Rhabdovirus genome reference strain 8743THA with the Rabies virus strain TH 13-42-07240 (TH2 clusters) and performed using Clustal Omega v1.2.0 [13] with default parameters. The DNA sequence variations between the Rabies virus strain 13-42-07240 sequences and the reference sequence (EU293111) were analyzed with the DnaSP software (version 5.10.1) to identify synonymous and non-synonymous sites.
Phylogenetic Analysis:
The trimmed original sequences were aligned with all Rabies virus sequences available from GenBank (May 2019) using Multiple Alignment of Fast Fourier Transform (MAFFT) [14]. Preliminary phylogenetic analyses were done using PhyML (maximum likelihood), implemented in Seaview Version 4, a multiplatform graphical user interface for sequence alignment and phylogenetic tree building [15]. Main phylogenetic analysis were performed on a subset of sequences, representing the overall genetic diversity of Rabies virus, under a Bayesian statistical framework implemented with BEAST package v.1.8.4 [16] using the model that fits best to the data accordingly with the corrected Bayesian Information Criterion (BICc) obtained in Jmodeltest2® [17].
RESULTS
Following the WHO’s recommendation, Rabies vaccine virus strain candidate should have a titer ≥6 log10 TCID50 (WHO, 2005). From the fifty rabies virus selected strains, after more than five passages in MNA cell and then passaged five to ten times in Vero cell line, only one strain, the Rabies virus strain TH 13-42-07240, was found positive by IFAT (4+, 100%), 1/50 (2%) and replicates efficiently at first passage in Vero cell (Figure 1) and after the 6th passages, it showed a titer of 6 log10 TCID50 on day 4 (MOI 0.1) and 6.2 log10 TCID50 on day 3 (MOI 0.01) (Table1).
|
Day |
Rabies virus strain TH 13-42-07240 |
|
|
Post-Infection |
MOI 0.1 |
MOI 0.01 |
|
0 |
3* |
2.1 |
|
1 |
3.6 |
2.8 |
|
2 |
4.6 |
4.3 |
|
3 |
5.6 |
6.2 |
|
4 |
6 |
5.6 |
|
5 |
5.8 |
5.4 |
|
6 |
5.5 |
5.3 |
|
7 |
5 |
5 |
Table 1: Replication kinetic of the Rabies virus strain TH 13-42-07240 from Thailand
Altogether, 251 strain sequences were used for phylogenic analysis of the G1 nucleoprotein gene and compared to the rabies virus strain TH 13-42-07240 (Figure F1).
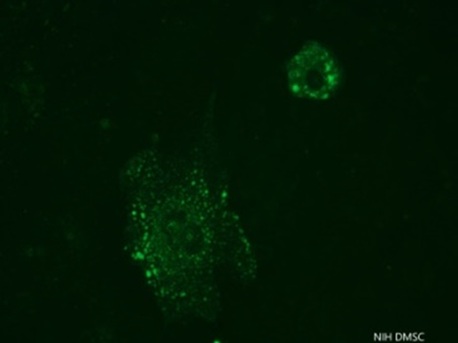 Figure 1: Indirect Immuno-Fluorescent Test on Vero cells monolayer infected by the Rabies virus strain TH 13-42-07240 adapted to Vero ATCC ® CCL-81 cell line
Figure 1: Indirect Immuno-Fluorescent Test on Vero cells monolayer infected by the Rabies virus strain TH 13-42-07240 adapted to Vero ATCC ® CCL-81 cell line
Caption: the observed granular plasmatic fluorescence is characteristic of the monoclonal antibody used for specifically binding to the nucleoprotein product of the virus.
Also, the rabies virus strain TH 13-42-07240 was different from the commonly use Rabies virus strain 13-42-07240 vaccine (Table 2, Figure 2), the TH 13-42-07240 rabies virus strain isolated from a canine brain appears widely distributed throughout Thailand (Table S1).
|
Gen Bank Acc. no. |
Reference no. |
Species |
Origin |
Year |
|
EU293121 |
8743THA |
Human |
Thailand |
1983 |
|
EU293111 |
8764THA |
Human |
Thailand |
1983 |
|
EU293115 |
9147FRA |
Fox |
France |
1991 |
|
EU293113 |
9001FRA |
Dog |
French Guyana |
1990 |
|
EU293116 |
9704ARG |
Bat |
Argentina |
1997 |
|
AY705373 |
SHBRV-18 |
Bat |
USA |
1983 |
|
EF437215 |
NNV-RAB-H |
Human |
India |
2006 |
|
EF206709 |
SAD B19 |
Vaccine |
- |
- |
|
NC_001542 |
PV |
Vaccine |
France |
1882 |
|
HQ317918 |
CTN-1-31 |
Human |
China |
1956 |
|
GQ918139 |
CVS-11 |
- |
France |
1882 |
|
EF206707 |
ERA |
Vaccine |
- |
- |
|
AB085828 |
HEP-Flury |
Vaccine |
USA |
1939 |
|
AY956319 |
Serotype 1 |
Saliva |
Germany |
- |
|
MN075931 |
TH13-42-07240 |
Dog |
Thailand |
1989 |
Table 2: Complete genome sequences of rabies virus used for genetic analysis
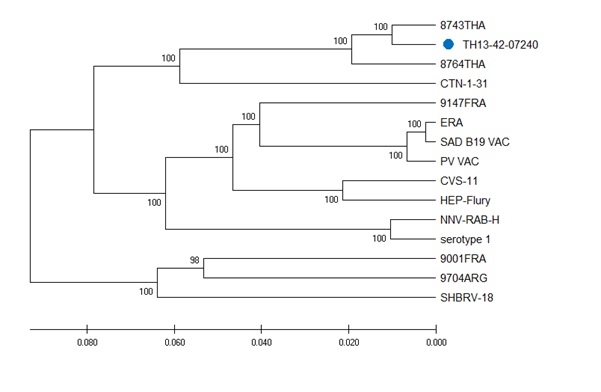 Figure 2: ML phylogeny of 15 selected Rabies virus strain sequences of the complete genome
Figure 2: ML phylogeny of 15 selected Rabies virus strain sequences of the complete genome
Legend: Blue circle = the selected vaccine candidate strain TH 13 -42-07240
DISCUSSION
Our study, with respect to the animal, use for Rabies virus amplification for diagnosis appears clearly unnecessary when for the first time NMA cell line were used to efficiently amplify Rabies virus followed by its adaptation to Vero cell culture. With respect to the TH 13-42-07240 rabies virus strain characteristics, a Rabies virus strain comply with WHO recommendation (i.e Vero cell line optimal titer, and availability of the companion IFA test) appears to be a strong candidate for the development of inactivated rabies vaccine that should efficiently stimulated immunity to prevent rabies epidemic in Thailand [21,22]. Furthermore, the rabies virus strain TH 13-42-07240 appears to actively circulate in Thailand and China and has the potential to be used for Rabies vaccine development of interest for the region [18]. Although, it has some genetic distance with the other commercial vaccine in the market, as a local strain isolated in Thailand it has been proven to efficiently by neutralizing circulated rabies virus strain [19,20].
CONCLUSION
Rabies virus strain TH 13-42-07240 originally isolated from Thailand, comply with international recommendation for Rabies candidate vaccine development, it is one of the most abundant strains in the South Eastern Asian peninsula, and therefore appears as a preferred candidate for Rabies vaccine development in Thailand.
ACKNOWLEDGEMENT
Funding for this research study was provided by National Vaccine Institute, Ministry of Public Health. Thank you for staff of Center for Vaccine Development, Institute of Molecular Biosciences, Mahidol University at Salaya and National Institute of Health, Thailand Department of Medical Sciences, Ministry of Public Health. We want to express our gratitude to Ms. Souki Gonzalez for writing assistance, technical and language editing.
REFERENCES
- Lu XX, Zhu WY, Wu GZ (2018) Rabies virus transmission via solid organs or tissue allotransplantation. Infect Dis Poverty 7:82.
- Dietzgen RG, Kondo H, Goodin MM, Kurath G, Vasilakis N (2017) The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res 227:158-70.
- Knobel DL, Cleaveland S, Coleman PG, Fevre EM, Meltzer MI (2005) Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organ 83:360–368.
- Jemberu WT, Molla W, Almaw G, Alemu S (2013) Incidence of rabies in humans and domestic animals and people's awareness in North Gondar Zone, Ethiopia. PLoS Negl Trop Dis 7:5
- Mitmoonpitak C, Wilde H, Tepsumetanon W (1997) Current status of animal rabies in Thailand. J Vet Med Sci 59: 457-460.
- Janchai (2016) Rabies and Situation in Thailand; 2018. Region 4-5 medical journal 2018; 37. Laboratory technique in rabies, 4th edition, World Health Organization, Geneva.
- Weekly Epidemiological Surveillance Report, Thailand. Bureau of Epidemiology, Department of Disease Control, Ministry of Public Health, 2019: 50.
- WHO Expert Committee on Rabies. World Health Organization Technical Report Series 709, World Health Organization, Geneva 1984.
- Meslin F.X, Kaplan M.H, Koprowski H (1996) Routine Laboratory Procedure in Laboratory Techniques in Rabies 4th Geneva. World Health Organization 55-122.
- Kamolvarin N, Tirawatnpong T, Rattanasiwamoke R, Tirawatnpong S, Panpanich T et al (1993) Diagnosis of rabies by polymerase chain reaction with nested primers. J Infect Dis 167:207-210.
- Narong Nitatpattana ,Yves Moné , Meriadeg AR Gouilh, Kumchol Chaiyo, Yutthana Joyjinda et al (2011) Genetic Diversity of Dengue-4 Virus Strains Isolated from Patients During a Single Outbreak of Dengue Fever, Thailand. J Fever 2:1009.
- Delmas O, Holmes EC, Talbi C, Larrous F, Dacheux L et al (2008) Genomic diversity and evolution of the Lyssaviruses.
- Tesh RB (1979) A method for the isolation and identification of dengue viruses, using mosquito cell cultures. Am J Tropical Med Hygiene 1053-1059.
- Boheemen S, De Graaf M, Lauber C, Bestebroer TM, Raj VS, et al (2012) Genomic Characterization of a Newly Discovered Coronavirus Associated with Acute Respiratory Distress Syndrome in Humans. mBio 3.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539.
- Katoh K, Standley DM (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol 30: 772-780.
- Gouy M, Guindon S, Gascuel O (2010) SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol Biol Evol 27: 221-224.
- Denduangboripant J, Wacharapluesadee S, Lumlertdacha B, Ruankaew N, Hoonsuwan W et al (2005) Transmission dynamics of rabies virus in Thailand: Implications for disease control. BMC Infection Diseases 5:52.
- Aga AM, Mekonnen Y, Hurisa B, Tesfaye T, Lemma H, et al.(2014) In vivo and in vitro cross neutralization studies of local rabies virus isolates with ERA based cell culture anti-rabies vaccine produced in Ethiopia. J Vaccines 5:256.
- World Health Organization, WHO Technical Report Series 931, WHO 2005; Geneva. 1–87.
- Minamoto N, Tanaka H, Hoshida M, Goto H, H Ito et al (1994) Linear and conformation-dependent antigenic sites on the nucleoprotein of rabies virus. Microbiol. Immunol 38:449-455
- Chan BK, Wilson T, Fischer KF, Kriesel JD. (2014) Deep sequencing to identify the cause of viral encephalitis.
SUPPLEMENTARY MATERIAL
|
|
Isolation no. |
Species |
Origin |
Year |
|
AY849022 |
5NBm |
Dog |
Nonthaburi |
2001 |
|
AY849023 |
26NPpmt |
Dog |
Nakhon Pathom |
2001 |
|
AY849024 |
53SPppd |
Dog |
Samut Prakan |
1999 |
|
AY849025 |
67PTtyb |
Dog |
Pathum Thani |
1999 |
|
AY849026 |
80PTllk |
Dog |
Pathum Thani |
1999 |
|
AY849027 |
81NBtn |
Dog |
Nonthaburi |
2001 |
|
AY849028 |
99PTkl |
Dog |
Pathum Thani |
1999 |
|
AY849029 |
79PTm |
Dog |
Pathum Thani |
1999 |
|
AY849030 |
125SSktb |
Dog |
Samut Sakhon |
1999 |
|
AY849031 |
133SSm |
Dog |
Samut Sakhon |
1999 |
|
AY849032 |
176SSktb |
Dog |
Samut Sakhon |
1999 |
|
AY849033 |
187SSm |
Dog |
Samut Sakhon |
1999 |
|
AY849034 |
207SSm |
Dog |
Samut Sakhon |
1999 |
|
AY849035 |
217SSm |
Dog |
Samut Sakhon |
1999 |
|
AY849036 |
218NBbk |
Dog |
Nonthaburi |
2001 |
|
AY849037 |
222NBbbt |
Dog |
Nonthaburi |
1999 |
|
AY849038 |
228NBtn |
Dog |
Nonthaburi |
1999 |
|
AY849039 |
304NPbl |
Dog |
Nakhon Pathom |
2001 |
|
AY849040 |
315NPm |
Dog |
Nakhon Pathom |
2001 |
|
AY849041 |
324NPm |
Dog |
Nakhon Pathom |
2001 |
|
AY849042 |
91PTns |
Dog |
Pathum Thani |
1999 |
|
AY849043 |
95NBtn |
Dog |
Nonthaburi |
1999 |
|
AY849044 |
108PTllk |
Dog |
Pathum Thani |
1999 |
|
AY849045 |
112PTtyb |
Dog |
Pathum Thani |
1999 |
|
AY849046 |
195NBby |
Dog |
Nonthaburi |
1999 |
|
AY849047 |
215NPncs |
Dog |
Nakhon Pathom |
1999 |
|
AY849048 |
232NPsp |
Dog |
Nakhon Pathom |
1999 |
|
AY849049 |
235NBpk |
Dog |
Nonthaburi |
1999 |
|
AY849050 |
263NPm |
Dog |
Nakhon Pathom |
1999 |
|
AY849051 |
C267BKbkn |
Cat |
Bangkok |
2000 |
|
AY849052 |
C269PTm |
Cat |
Pathum Thani |
2000 |
|
AY849053 |
C271BKrtv |
Cat |
Bangkok |
2001 |
|
AY849054 |
C274BKdd |
Cat |
Bangkok |
1999 |
|
AY849055 |
C276BKpv |
Cat |
Bangkok |
1999 |
|
AY849056 |
C277BKkt |
Cat |
Bangkok |
1999 |
|
AY849057 |
303KJtmk |
Dog |
Kanchanaburi |
2001 |
|
AY849058 |
308KJm |
Dog |
Kanchanaburi |
2001 |
|
AY849059 |
318KJtmk |
Dog |
Kanchanaburi |
2001 |
|
AY849060 |
326KJtm |
Dog |
Kanchanaburi |
2001 |
|
AY849061 |
333KJm |
Dog |
Kanchanaburi |
2001 |
|
AY849062 |
335KJpnt |
Dog |
Kanchanaburi |
2001 |
|
AY849063 |
89PTtb |
Dog |
Pathum Thani |
1999 |
|
AY849064 |
38/43 |
Dog |
Chaiyaphum |
2000 |
|
AY849065 |
39/43 |
Dog |
Chaiyaphum |
2000 |
|
AY849066 |
46/43 |
Dog |
Chaiyaphum |
2000 |
|
AY849067 |
23SPppd |
Dog |
Samut Prakan |
2001 |
|
AY849068 |
33/43 |
Dog |
Chaiyaphum |
2000 |
|
AY849069 |
34/43 |
Dog |
Chaiyaphum |
2000 |
|
AY849070 |
19/43 |
Dog |
Chaiyaphum |
2000 |
|
AY849071 |
51BKds |
Dog |
Bangkok |
1999 |
|
AY849072 |
62SPbp |
Dog |
Samut Prakan |
1999 |
|
AY849073 |
48BKpyt |
Dog |
Bangkok |
2001 |
|
AY849074 |
HM65BK |
Human |
Bangkok |
1998 |
|
AY849075 |
HM75BK |
Human |
Bangkok |
1998 |
|
AY849076 |
HM88BKjj |
Human |
Bangkok |
1999 |
|
AY849077 |
HM208BKpv |
Human |
Bangkok |
2001 |
|
AY849078 |
404PLkcs |
Cattle |
Phatthalung |
2002 |
|
AY849079 |
355UThk |
Dog |
Uthai Thani |
2001 |
|
AY849080 |
415PLtm |
Dog |
Phatthalung |
2002 |
|
AY849081 |
294CPm |
Dog |
Chaiyaphum |
2002 |
|
AY849082 |
295CPksb3 |
Dog |
Chaiyaphum |
2002 |
|
AY849083 |
351NSm |
Dog |
Nakhon Sawan |
2001 |
|
AY849084 |
353CNmnr |
Dog |
Chai Nat |
2001 |
|
|
363NScs |
Dog |
Nakhon Sawan |
2001 |
|
AY849086 |
412SKsd |
Dog |
Songkla |
2002 |
|
AY849087 |
414SKm |
Dog |
Songkla |
2002 |
|
AY849088 |
361NSly |
Dog |
Nakhon Sawan |
2001 |
|
AY849089 |
376SHib |
Dog |
Sing Buri |
2001 |
|
AY849090 |
380UTbr |
Dog |
Uthai Thani |
2001 |
|
AY849091 |
393LBm |
Dog |
Lop Buri |
2001 |
|
AY849092 |
384SHib |
Dog |
Sing Buri |
2001 |
|
AY849093 |
411NTht |
Dog |
Nakhon Si Thammarat |
2002 |
|
AY849094 |
374CNspy |
Dog |
Chai Nat |
2001 |
|
AY849095 |
362CNm |
Dog |
Chai Nat |
2001 |
|
AY849096 |
357UTm |
Dog |
Uthai Thani |
2001 |
|
AY849097 |
356UTth |
Dog |
Uthai Thani |
2001 |
|
AY849098 |
381NSttk |
Dog |
Nakhon Sawan |
2001 |
|
AY849099 |
408SKhy |
Dog |
Songkla |
2002 |
|
AY849100 |
332SBm |
Dog |
Suphan Buri |
2001 |
|
AY849101 |
354CNhk |
Dog |
Chai Nat |
2001 |
|
AY849102 |
425PN |
Dog |
Phisanulok |
2002 |
|
AY849103 |
389LBm |
Dog |
Lop Buri |
2001 |
|
AY849104 |
423PN |
Dog |
Phisanulok |
2002 |
|
AY849105 |
424PN |
Dog |
Phisanulok |
2002 |
|
AY849106 |
413SKhy |
Cattle |
Songkla |
2001 |
|
AY849107 |
358CNsbr |
Dog |
Chai Nat |
2001 |
|
AY849108 |
317SBspn |
Dog |
Suphan Buri |
2001 |
|
AY849109 |
334PJsry |
Dog |
Prachuap Khiri Khan |
2001 |
|
AY849110 |
340SBspn |
Dog |
Suphan Buri |
2001 |
|
AY849111 |
400SKm |
Dog |
Songkla |
2001 |
|
AY849112 |
87BKsl |
Dog |
Bangkok |
2001 |
|
AY849113 |
156PTns |
Dog |
Pathum Thani |
1999 |
|
AY849114 |
182CCbnp |
Dog |
Chachoengsao |
1999 |
|
AY849115 |
349PBm |
Dog |
Phetchaburi |
2001 |
|
AY849116 |
307RBptr |
Dog |
Ratchaburi |
2001 |
|
AY849117 |
329PBm |
Dog |
Phetchaburi |
2001 |
|
AY849118 |
406STm |
Dog |
Satun |
2002 |
|
AY849119 |
352NStk |
Dog |
Nakhon Sawan |
2001 |
|
AY849120 |
301RBm |
Dog |
Ratchaburi |
2001 |
|
AY849121 |
157PJsry |
Dog |
Prachuap Khiri Khan |
1999 |
|
AY849122 |
237NYm |
Dog |
Nakhon Nayok |
1999 |
|
AY849123 |
270ATvsc |
Cat |
Ang Thong |
2001 |
|
AY849124 |
281NRm |
Dog |
Nakhon Ratchasima |
2001 |
|
AY849125 |
282NRpc |
Dog |
Nakhon Ratchasima |
2001 |
|
AY849126 |
305RBbp |
Dog |
Ratchaburi |
2001 |
|
AY849127 |
306RBptr |
Dog |
Ratchaburi |
2001 |
|
AY849128 |
191AYsn |
Dog |
Ayutthaya |
1999 |
|
AY849129 |
151SBdc |
Dog |
Suphan Buri |
1999 |
|
AY849130 |
22CBkhm |
Dog |
Chanthaburi |
2001 |
|
AY849131 |
162PCm |
Dog |
Phichit |
1999 |
|
AY849132 |
319PBm |
Dog |
Phetchaburi |
2001 |
|
AY849133 |
86SPm |
Dog |
Samut Prakan |
2001 |
|
AY849134 |
136CLblm |
Dog |
Chon Buri |
1999 |
|
AY849135 |
396YLm |
Dog |
Yala |
2001 |
|
AY849136 |
250AYbt |
Dog |
Ayutthaya |
1999 |
|
AY849137 |
302RBpt |
Dog |
Ratchaburi |
2001 |
|
AY849138 |
316SMm |
Dog |
Samut Songkhram |
2001 |
|
AY849139 |
426PN |
Dog |
Phisanulok |
2002 |
|
AY849140 |
296CPksb |
Dog |
Chaiyaphum |
2002 |
|
AY849141 |
313PBnyp |
Dog |
Phetchaburi |
2001 |
|
AY849142 |
288NRbl |
Dog |
Nakhon Ratchasima |
2002 |
|
AY849143 |
505KBlt |
Dog |
Krabi |
2000 |
|
AY849144 |
507TRrd |
Dog |
Trang |
2000 |
|
AY849145 |
511NTts |
Dog |
Nakhon Si Thammarat |
2000 |
|
AY849146 |
515KBlt |
Dog |
Krabi |
2000 |
|
AY849147 |
524KBkn |
Dog |
Krabi |
2000 |
|
AY849148 |
559TRpl |
Cattle |
Trang |
2000 |
|
AY849149 |
578KBkt |
Dog |
Krabi |
2001 |
|
AY849150 |
584TRkt |
Dog |
Satun |
2001 |
|
AY849151 |
603KBlt |
Dog |
Krabi |
2001 |
|
AY849152 |
595Ylbns |
Dog |
Yala |
2001 |
|
AY849153 |
493RNm |
Dog |
Ranong |
2000 |
|
AY849154 |
656PLppy |
Dog |
Phatthalung |
2002 |
|
AY849155 |
676KSm |
Squirrel |
Kalasin |
2002 |
|
AY849156 |
513PLm |
Dog |
Phatthalung |
2000 |
|
AY849157 |
589YLm |
Dog |
Yala |
2001 |
|
AY849158 |
608SRm |
Dog |
Satun |
2001 |
|
AY849159 |
HMS152S |
Human |
Si Sa Ket |
2001 |
|
AY849160 |
599TRhy |
Dog |
Trang |
2001 |
|
AY849161 |
473BRpk |
Dog |
Surin |
2001 |
|
AY849162 |
485BRpk |
Dog |
Buri Ram |
2002 |
|
AY849163 |
510NTm |
Dog |
Nakhon Si Thammarat |
2000 |
|
AY849164 |
486UMhsp |
Dog |
Amnat Chareon |
2002 |
|
AY849165 |
487Umm |
Dog |
Amnat Charoen |
2002 |
|
AY849166 |
501RNm |
Dog |
Nakhon Si Thammarat |
2002 |
|
AY849167 |
283NRpm |
Dog |
Nakhon Ratchasima |
2001 |
|
AY849168 |
459KSm |
Dog |
Kalasin |
2002 |
|
AY849169 |
463SRsn |
Dog |
Surin |
2001 |
|
AY849170 |
464SRskp |
Dog |
Surin |
2001 |
|
AY849171 |
465SRm |
Dog |
Surin |
2001 |
|
AY849172 |
466SRrbr |
Dog |
Surin |
2001 |
|
AY849173 |
528NTrpb |
Dog |
Nakhon Si Thammarat |
2000 |
|
AY849174 |
548NTlsk |
Dog |
Nakhon Si Thammarat |
2000 |
|
AY849175 |
289NRht |
Dog |
Nakhon Ratchasima |
2001 |
|
AY849176 |
723KKm |
Dog |
Khon Kaen |
2001 |
|
AY849177 |
481BRhr |
Dog |
Buri Ram |
2002 |
|
AY849178 |
494RNm |
Dog |
Ranong |
2000 |
|
AY849179 |
503SNws |
Dog |
Surat Thani |
2002 |
|
AY849180 |
553STdkl |
Dog |
Khuan Ka Long/Satun |
2000 |
|
AY849181 |
666STm |
Cattle |
Satun |
2002 |
|
AY849182 |
458Lam |
Dog |
RoiEt |
2001 |
|
AY849183 |
500SNks |
Dog |
Surat Thani |
2002 |
|
AY849184 |
454Lam |
Dog |
Roi Et |
2001 |
|
AY849185 |
495RNm |
Dog |
Ranong |
2001 |
|
AY849186 |
460Lam |
Dog |
Roi Et |
2002 |
|
AY849187 |
488Umm |
Dog |
Amnat Charoen |
1998 |
|
AY849188 |
489SEkh |
Dog |
Si Sa Ket |
2000 |
|
AY849190 |
502SNcb |
Dog |
Surat Thani |
2002 |
|
AY849189 |
472BRm |
Dog |
Buri Ram |
2001 |
|
AY849191 |
499SNps |
Dog |
Surat Thani |
2002 |
|
AY849192 |
HMS241CL |
Human |
Chon Buri |
2001 |
|
AY849193 |
694LAsp |
Dog |
Roi Et |
2002 |
|
AY849194 |
690NKsps |
Dog |
Nong Khai |
2002 |
|
AY849195 |
689Umm |
Dog |
Amnat Charoen |
2001 |
|
AY849196 |
688Umm |
Dog |
Amnat Charoen |
2002 |
|
AY849197 |
691NKbk |
Dog |
Nong Khai |
2002 |
|
AY849198 |
695Lack |
Dog |
Roi Et |
2002 |
|
AY849199 |
698KSm |
Squirrel |
Kalasin |
2002 |
|
AY849200 |
700KStkt |
Dog |
Kalasin |
2003 |
|
AY849201 |
708YSm |
Dog |
Yasothon |
2000 |
|
AY849202 |
568YLrm |
Dog |
Yala |
2000 |
|
AY849203 |
725MDm |
Dog |
Mukdahan |
2001 |
|
AY849204 |
726UBbt |
Dog |
Ubol Ratchathani |
2001 |
|
AY849205 |
738KKm |
Dog |
Khon Kaen |
2002 |
|
AY849206 |
742MDm |
Dog |
Mukdahan |
2002 |
|
AY849207 |
740MDdt |
Dog |
Mukdahan |
2002 |
|
AY849208 |
711SLm |
Dog |
Sakon Nakhon |
2000 |
|
AY849209 |
709SEktr |
Dog |
Si Sa Ket |
2000 |
|
AY849210 |
678cBRm |
Cat |
Buri Ram |
2000 |
|
AY849211 |
728UBm |
Cattle |
Ubol Ratchathani |
2001 |
|
AY849212 |
685BRppc |
Dog |
Buri Ram |
2002 |
|
AY849213 |
714YSm |
Dog |
Yasothon |
2001 |
|
AY849214 |
715YSm |
Water buffalo |
Yasothon |
2001 |
|
AY849215 |
718YSlkk |
Dog |
Yasothon |
2001 |
|
AY849216 |
705MDm |
Dog |
Mukdahan |
2000 |
|
AY849217 |
731UBvrc |
Dog |
Ubol |
2001 |
|
AY849218 |
704KKcp |
Dog |
Khon Kaen |
2000 |
|
AY849219 |
703KKm |
Dog |
Khon Kaen |
2000 |
|
AY849220 |
732KSm |
Cattle |
Kalasin |
2001 |
|
AY849221 |
741YSm |
Dog |
Yasothon |
2002 |
|
AY849222 |
744UBdud |
Dog |
Ubol Ratchathani |
2002 |
|
AY849223 |
713KKcp |
Dog |
Khon Kaen |
2001 |
|
AY849224 |
717UBm |
Dog |
Ubol Ratchathani |
2001 |
|
AY849225 |
707LYm |
Dog |
Loei |
2000 |
|
AY849226 |
131SPpsj |
Dog |
Samut Prakan |
1999 |
|
AY849227 |
HMS223RY |
Human |
Rayong |
2002 |
|
AY849228 |
747PGtp |
Dog |
Phangnga |
2003 |
|
AY849229 |
766PRm |
Dog |
Phetchabun |
2003 |
|
AY849230 |
769PRcd |
Dog |
Phetchabun |
2003 |
|
AY849232 |
774NNm |
Dog |
Nan |
2002 |
|
AY849231 |
773CMcp |
Dog |
Chiang Mai |
2002 |
|
AY849233 |
775PYp |
Dog |
Phayao |
2003 |
|
AY849234 |
776NStk |
Dog |
Nakhon Sawan |
2002 |
|
AY849235 |
795NScs |
Dog |
Nakhon Sawan |
2003 |
|
AY849236 |
801SUsk |
Dog |
Sukhothai |
2002 |
|
AY849237 |
802KPlkb |
Dog |
Kamphaeng Phet |
2002 |
|
AY849238 |
811SUkrm |
Dog |
Sukhothai |
2002 |
|
AY849239 |
806PNnm |
Dog |
Phisanulok |
2002 |
|
AY849240 |
814UDm |
Dog |
Uttaradit |
2002 |
|
AY849241 |
807UDts |
Dog |
Uttaradit |
2002 |
|
AY849242 |
800Udm |
Dog |
Uttaradit |
2002 |
|
AY849243 |
815PRls |
Dog |
Phetchabun |
2002 |
|
AY849244 |
805PCbmn |
Dog |
Phichit |
2002 |
|
AY849245 |
780NSm |
Dog |
Nakhon Sawan |
2003 |
|
AY849246 |
762PRm |
Dog |
Phetchabun |
2003 |
|
AY849247 |
813PNbrk |
Dog |
Phisanulok |
2002 |
|
AY849248 |
796PNbrk |
Dog |
Phisanulok |
2003 |
|
AY849249 |
816PRcd |
Dog |
Phetchabun |
2002 |
|
AY849250 |
778LBtv |
Dog |
Lop Buri |
2003 |
|
AY849251 |
794SUssr |
Dog |
Sukhothai |
2003 |
|
AY849252 |
784Sum |
Dog |
Sukhothai |
2003 |
|
AY849253 |
785SUkkl |
Dog |
Sukhothai |
2003 |
|
AY849254 |
788SUsk |
Dog |
Sukhothai |
2003 |
|
AY849255 |
808SUssn |
Dog |
Sukhothai |
2002 |
|
AY849256 |
793SUsn |
Dog |
Sukhothai |
2003 |
|
AY849257 |
777LBm |
Dog |
Lop Buri |
2003 |
|
AY849258 |
779NSly |
Dog |
Nakhon Sawan |
2003 |
|
AY849259 |
804UDm |
Dog |
Uttaradit |
2002 |
|
AY849260 |
338PJm |
Dog |
Prachuap Khiri Khan |
2001 |
|
HQ317918 |
CTN-1-31 |
Human |
China |
1956 |
|
EU293115 |
9147FRA |
Fox |
France |
1991 |
|
EU293113 |
9001FRA |
Dog |
French Guyana |
1990 |
|
AY705373 |
SHBRV-18 |
Bat |
USA |
1983 |
|
EU293116 |
9704ARG |
Bat |
Argentina |
1997 |
|
EF206709 |
SAD B19 |
Vaccine |
- |
- |
|
EF206707 |
ERA |
Vaccine |
- |
- |
|
NC_001542 |
PV |
Vaccine |
France |
1882 |
|
AB085828 |
HEP-Flury |
Vaccine |
USA |
1939 |
|
GQ918139 |
CVS-11 |
- |
France |
1882 |
|
EF437215 |
NNV-RAB-H |
Human |
India |
2006 |
|
AY956319 |
Serotype 1 |
Saliva |
Germany |
- |
|
MN075931 |
TH13-42-07240 |
Dog |
Thailand |
1989 |
Table S1: Nucleoprotein Gene sequences of the selected rabies virus strains used for genetic analysis
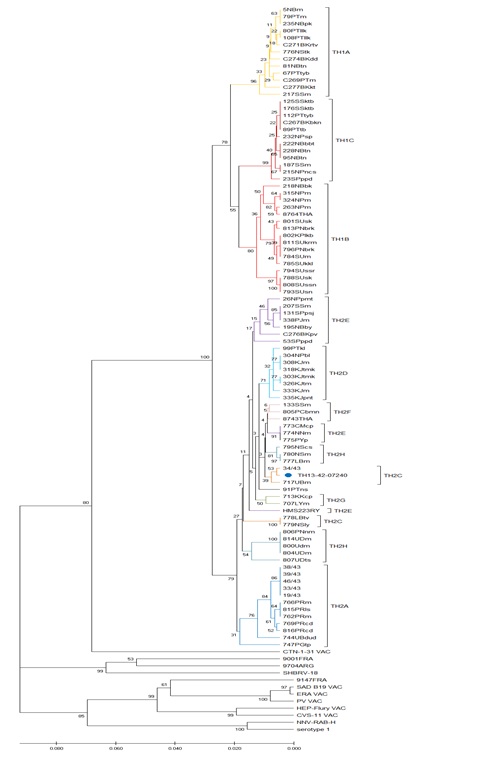 Figure S1: ML phylogeny of xxx sequences from the N-coding region of Rabies virus strains.
Figure S1: ML phylogeny of xxx sequences from the N-coding region of Rabies virus strains.
Caption: Sequences were obtained from Gen Bank and the TH strain sequenced for this study.
Citation: Nitatpattana N, Primsirikunawut A, Buree S, Chuapudee D, Rodpai E, Chaiyo K, Chansiprasert K, Nakgoi K, Poolam K, Lukebuo A, Sangkitporn S,Mandja BM, Gonzalez JP (2020) Selection of a Rabies Virus Strain as a New Candidate for Vaccine Development in Thailand. J Vaccines Res Vaccin , 6: 015.
Copyright: © 2020 Nitatpattana N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

