Transcriptomic Regulation of Lung Lavage Cells from Pulmonary Fibrosis Patients under the Inhibition of PI3K
*Corresponding Author(s):
Mengqiao WangDepartment Of Epidemiology And Biostatistics, West China School Of Public Health And West China Fourth Hospital, Sichuan University, Chengdu, Sichuan Province, China
Tel:+86 13880649045,
Email:mengqiaowang@gmail.com
Abstract
Pneumoconiosis refers to a series of lung diseases caused by inhalation of mineral dust and the main pathological characteristics are chronic lung inflammation and progressive pulmonary fibrosis. This study aims to analyze the changes in gene expression and cytological function of cells upon the inhibition of PI3K. Wortmannin was used to specifically inhibit PI3K. Transcriptomics methods were used to compare the Differentially Expressed Genes (DEGs) between the inhibitory group and the control group. The Protein-Protein Interaction (PPI) network of DEGs was constructed and the PI3K pathway was demonstrated with the use of the Oncomine database. Inhibiting PI3K of lung lavage cells (e.g. alveolar macrophages) from pneumoconiosis patients can cause a spectrum of changes in cellular functions and pathways, thus affecting the pulmonary fibrosis process within patients. This study provides patient cell-based understanding of the molecular mechanisms of pneumoconiosis and reveal potential molecular targets and diagnostic biomarkers for the treatment of pneumoconiosis.
Keywords
Alveolar macrophages; Pulmonary fibrosis; PI3K signaling pathway
Methods
Whole Lung Lavage (WLL)
Patients diagnosed of pulmonary fibrosis (pneumoconiosis) received WLL at certified clinical institutes as the recommended medical treatment. During WLL, 0.9% normal saline solution was heated to 37°C, subjected to lung lavage 20 minutes after unilateral pure oxygen ventilation, and terminated when the drainage fluids turn from turbid to fully transparent. The total volume of lung lavage fluids, routinely discarded as medical wastes, was generally 4000-6000 ml, influenced by the patients’ lung volume and pneumoconiosis stages. Patients were informed of the proposed research and gave permission to authorize the usage of their lung lavage fluids for biomedical research purposes. Accordingly, after WLL, lung lavage fluids collected into the aseptic containers were not discarded but immediately transferred to the laboratory for further biomedical experiments. The study design was reviewed and approved by the institution’s research ethics committee.
Cell isolation from lung lavage fluids
The lung lavage fluid was filtered through double-layer gauze to remove mucus and other clumped stuffs, aliquoted to 50 ml tubes, and then centrifuged at 1800 rpm for 5 min. Supernatants were discarded and the pellets were washed 2-3 times with PBS. The pellets were aggregated and resuspended in culture medium, made up of DMEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1X penicillin-streptomycin mixture (HyClone). Cells were imaged under a light microscope to evaluate quality, measured for density and cultured in 25 cm2 plastic flasks (Corning) in an incubator (5% CO2, 37°C). For cryopreservation, cells were resuspended in culture medium supplemented with 10% dimethyl sulfoxide (Sigma-Aldrich) and frozen in stock tubes at a -80°C freezer.
Inhibition treatment and design
Cells were incubated at 37°C in a humidified 5% CO2 incubator and divided into a control group and a treatment group of PI3K inhibition by wortmannin. Cells in the control group were grown in the culture medium and cells in the experiment group were grown in the culture medium supplemented with 10 nM wortmannin for 6h. Both control and experiment groups were repeated with independent triplicates.
RNA-seq experiment and bioinformatic analysis
Total RNA was extracted from cells and mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation of mRNA was carried out using divalent cations under elevated temperature, followed by cDNA synthesis using random hexamer primers. After adenylation of 3’-end DNA fragments, adaptors with hairpin loop structure were ligated to prepare for hybridization. The library fragments were purified with AMPure XP system (Beckman Coulter) to select cDNA fragments of about 400 bp in length. After PCR reactions, products were purified and the library quality was assessed on a Bioanalyzer 2100 system (Agilent). The library preparations were finally sequenced on a Novaseq platform (Illumina) with the sequence generation of 150 bp paired-end reads. All RNA-seq experiments were repeated in independent triplicates.
Raw reads in FASTQ formats were preprocessed, aligned to the reference genome (GRCh38) using Hisat2 and converted into BAM files for followed-up RNA-seq analysis [19,20] in R. Differential expression was based on the DESeq2 analysis pipeline, and the thresholds of absolute log2 (fold change) over 1 and adjusted P value less than 0.05 were used to identify Differentially Expressed Genes (DEGs).
Construction of Protein-Protein Interaction (PPI) network
The Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org/) [21], is an online tool designed to explore and analyze PPI information.In order to evaluate the interaction between DEGs after inhibition experiments, STRING is used to map DEGs, and the interaction with combination score > 0.7 is selected. The top 10 most connected genes in the network are defined as hub genes to identify key elements in Biological Processes (BP).
Verification of gene expression in Oncomine database
Oncomine (www.oncomine.org; Ion Torrent; Thermo Fisher Scientific, Inc.) is an online cancer microarray database that facilitates genome-wide expression analysis, within which we conducted analysis of PI3K inhibition in squamous cell lung carcinoma (SQCLC). Weiss, et al. [22], Talbot, et al. [23] and Bhattacharjee, et al. [24] lung cancer gene expression data in the Oncomine database were retrieved for expression levels of PI3K pathway with Pvalue
Statistical Analysis
Data analysis and visualization were performed in the R statistical software. For single null-hypothesis tests, a significance level of 0.05 was used. For multiple testing where the control of False Discovery Rate (FDR) was necessary, raw P values were adjusted by the Benjamini-Hochberg (BH) method to obtain adjusted P values.
Results
The main pathological features of pneumoconiosis are chronic pulmonary inflammation and progressive pulmonary fibrosis. The activation of immune response in lung microenvironment of patients with pulmonary fibrosis is related to the induction of PI3K pathway, but the molecular mechanism of pneumoconiosis induced pulmonary fibrosis has not been fully elucidated. Under the activation of the immune response in the lung microenvironment of patients with pulmonary fibrosis, lung lavage cells (mainly alveolar macrophages etc.) maintain their innate transcriptional profiles. Upon the inhibition of PI3K pathway by wortmannin, a spectrum of transcriptional changes is expected but unknown. Using RNA-seq as the next-generation high-throughput sequencing technology, such gene expression changes could be revealed by the profiles of Differentially Expressed Genes (DEGs). For the control treatment vs. the experimental treatment, three replicates were prepared and assayed in parallel. With parameter thresholds set as the absolute log2 fold change at more than 1 and the adjusted p-value smaller than 0.05 to control the error Discovery Rate (FDR), the volcano plot reveals the transcriptional changes of patient-derived lung lavage cells upon inhibition of PI3K by wortmannin (Figure 1). In comparison, with PI3K inhibition, a total of 3356 genes were identified as differentially expressed genes (1990 up-regulated genes and 1366 down-regulated genes). The MA plot demonstrated that the DEGs are predominantly highly expressed genes thus the observed differential expression could hardly be attributed to false discoveries due to low expression levels (Figure 2).
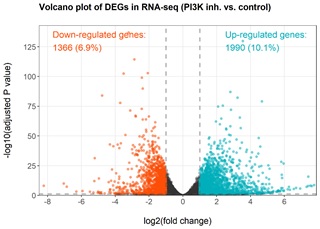 Figure 1: Volcanic plot of DEGs obtained after inhibiting PI3K target. A total of 3356 genes were identified as differentially expressed genes (1990 up-regulated genes and 1366 down-regulated genes). The up- and down-regulated genes before and after inhibition were annotated. Thresholds of absolute log2 (fold change) of 1 and adjusted P value of 0.05 were applied to identify DEGs.
Figure 1: Volcanic plot of DEGs obtained after inhibiting PI3K target. A total of 3356 genes were identified as differentially expressed genes (1990 up-regulated genes and 1366 down-regulated genes). The up- and down-regulated genes before and after inhibition were annotated. Thresholds of absolute log2 (fold change) of 1 and adjusted P value of 0.05 were applied to identify DEGs.
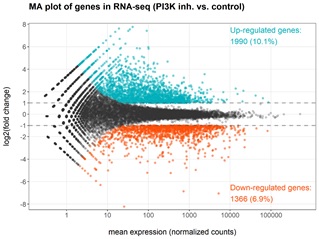 Figure 2: MA plot of DEGs obtained after inhibiting PI3K target. The DEGs are predominantly highly expressed genes thus the observed differential expression could hardly be attributed to false discoveries due to low expression levels.
Figure 2: MA plot of DEGs obtained after inhibiting PI3K target. The DEGs are predominantly highly expressed genes thus the observed differential expression could hardly be attributed to false discoveries due to low expression levels.
Upon inhibiting PI3K, 229 identified DEGs (log2 Fold Change > 3, Padj < 0.0001) were included in the analysis within the STRING online database. A total of 44 genes were filtered into the PPI network complex, including 217 nodes and 132 edges, and 185 genes were located outside the DEGs PPI network (Figure 3). Subsequently, 10 hub genes with the highest connectivity in the network were identified, which were Angiopoietin Like 4 (ANGPTL4), Ankyrin Repeat Domain 1 (ANKRD1), ATPase H+ transporting V1 subunit G2 (ATP6V1G2), Calmodulin Regulated Spectrin Associated Protein family member 3 (CAMSAP3), Coiled-Coil Domain Containing protein 151 (CCDC151), C-C motif Chemokine Ligand 1 (CCL1), C-C motif Chemokine Ligand 15 (CCL15), C-C motif Chemokine Ligand 2 (CCL2), C-C motif Chemokine Ligand 8 (CCL8) and chromatin licensing and DNA replication factor 1 (CDT1) (Figure 4). The top hub genes were evenly distributed regarding up-regulation or down-regulation so the underlining transcriptomic changes are bi-directional. The clustered heatmap of these 10 hub genes among the triplicate experiment samples reveals significant reproductivity and harmonized association (Figure 5). Discoveries of the DEGs and especially the top hub genes and their associated pathways identify the physiological and biochemical processes perturbed upon the inhibition of PI3K within lung lavage cells, suggesting the potential panel of genes for functional targeting.
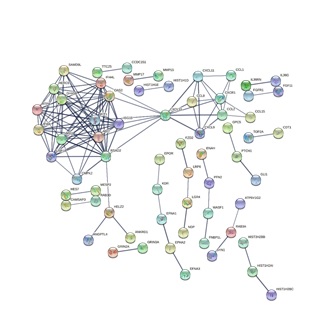 Figure 3: PPI network constructed from the DEGs and hub genes. Using the STRING online database, a total of 44 DEGs were filtered into the DEGs PPI network complex. The nodes represent proteins and the edges represent the interaction of proteins.
Figure 3: PPI network constructed from the DEGs and hub genes. Using the STRING online database, a total of 44 DEGs were filtered into the DEGs PPI network complex. The nodes represent proteins and the edges represent the interaction of proteins.
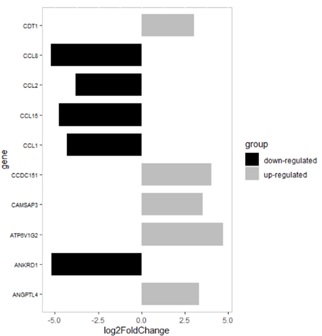 Figure 4: Expression level of the top 10 hub genes. 5 up-regulated hub genes are C-C motif chemokine ligand 1 (CCL1), C-C motif chemokine ligand 15 (CCL15), C-C motif chemokine ligand 2 (CCL2), C-C motif chemokine ligand 8 (CCL8) and ankyrin repeat domain 1 (ANKRD1); 5 down-regulated hub genes are angiopoietin like 4 (ANGPTL4), ATPase H+ transporting V1 subunit G2 (ATP6V1G2), calmodulin regulated spectrin associated protein family member 3 (CAMSAP3), Coiled-coil domain containing protein 151 (CCDC151)and chromatin licensing and DNA replication factor 1 (CDT1).
Figure 4: Expression level of the top 10 hub genes. 5 up-regulated hub genes are C-C motif chemokine ligand 1 (CCL1), C-C motif chemokine ligand 15 (CCL15), C-C motif chemokine ligand 2 (CCL2), C-C motif chemokine ligand 8 (CCL8) and ankyrin repeat domain 1 (ANKRD1); 5 down-regulated hub genes are angiopoietin like 4 (ANGPTL4), ATPase H+ transporting V1 subunit G2 (ATP6V1G2), calmodulin regulated spectrin associated protein family member 3 (CAMSAP3), Coiled-coil domain containing protein 151 (CCDC151)and chromatin licensing and DNA replication factor 1 (CDT1).
 Figure 5: Expression heatmap of the top 10 hub genes. Heatmap showing expression profiles of differentially expressed 10 hub genes between the control group and the PI3K-inhibition group.
Figure 5: Expression heatmap of the top 10 hub genes. Heatmap showing expression profiles of differentially expressed 10 hub genes between the control group and the PI3K-inhibition group.
Target genes of PI3K pathway were analyzed in the Oncomine database to explore the relationship between this signaling pathway and Squamous Cell Lung Carcinoma (SQCLC), a type of cancer that is at least partially related to pulmonary fibrosis. The mRNA expression levels of PI3K and its downstream effectors such as AKT and mTOR in SQCLC samples were significantly different from those in normal lung samples (Figure 6), suggesting PI3K and the downstream pathway is a key player in the pulmonary fibrosis within such patients.
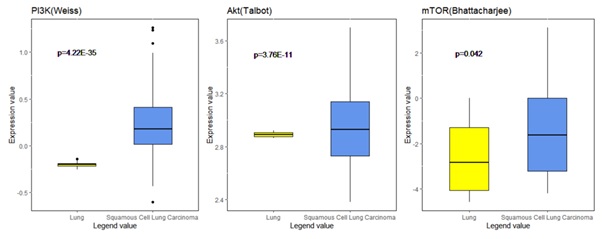 Figure 6: Analysis of expression of PI3K/Akt/mTOR signaling pathway in the Oncomine database. Gene expression data was obtained from Weiss, Talbot and Bhattacharjee lung datasets and analyzed with Oncomine. mRNA expression levels of PI3K, Akt and mTOR in normal lung vs. SQCLC were compared. Log-transformed expression levels are displayed as box plots.
Figure 6: Analysis of expression of PI3K/Akt/mTOR signaling pathway in the Oncomine database. Gene expression data was obtained from Weiss, Talbot and Bhattacharjee lung datasets and analyzed with Oncomine. mRNA expression levels of PI3K, Akt and mTOR in normal lung vs. SQCLC were compared. Log-transformed expression levels are displayed as box plots.
Discussion
To predict the functional association among the identified DEGs upon inhibition of PI3K in lung lavage cells, we constructed PPI networks for the inhibition comparison, and selected the top 10 hub genes with the highest connectivity for further analysis, including ANGPTL4, ANKRD1, ATP6V1G2, CAMSAP3, CCDC151, CCL1, CCL15, CCL2, CCL8 and CDT1, which were mainly related to the functions of immune system process, monocyte chemotaxis, regulation of leukocyte migration etc. Functions of individual genes are key to the understanding of their specific roles. For example, protein encoded by angiopoietin like 4 (ANGPTL4) regulates chronic inflammation, angiogenesis and vascular permeability [25], in which inflammation plays a key role in the progression of pneumoconiosis to pulmonary fibrosis [6]. Previous in vitro studies have shown that activation of mTOR in lung epithelial cells, a serine/threonine protein kinase in the PI3K-related kinase family, promotes pulmonary fibrosis and expression of ANGPTL4 [26]. In addition, four up-regulated hub genes (CCL1, CCL15, CCL2 and CCL8) are related to chemokines. C-C motif Chemokine Ligand 1 (CCL1) and C-C motif Chemokine Ligand 2 (CCL2) are cytokine genes clustered on the q arm of chromosome 17, leading to the production of chemokines involved in immune regulation and inflammation. Such cytokines have chemotactic activity on monocytes and basophils, but with little chemotactic activity on neutrophils and eosinophils. Animal experiments further demonstrated that the expression of CCL2 was up-regulated in acute and chronic silicosis models [27,28].
Among the downregulated hub genes, calmodulin regulated spectrin associated protein family member 3 (CAMSAP3) is required to maintain microtubule organization and cell morphogenesis [29]. Deletion of CAMSAP3 in human lung carcinoma-derived cell lines, leading to the upregulation of Akt proteins (also known as protein kinase B proteins, the downstream molecule of PI3K) activity [29]. Highly induced activation of PI3K can lead to multiple cancer characteristics, and this pathway is an attractive target for new anticancer therapy.One study found that in a large number of fibrotic silicosis, the incidence of SQCLC, a type of Non-Small Cell Lung Cancer (NSCLC), is higher [30]. Mutations in PI3K and its downstream effectors frequently occurred in NSCLC patients [31]. The molecular mechanism of the occurrence and development of pneumoconiosis-induced lung squamous cell carcinoma has not been fully elucidated, while our study suggests that CAMSAP3 may be involved in this process.Similarly, we demonstrated with the use of the Oncomine database that PI3K pathway was differentially expressed in SQCLC vs. normal lung tissues. Therefore, inhibiting the PI3K pathway of AMs may not only affect pulmonary fibrosis, but also affect SQCLC caused by pulmonary fibrosis.
Inhibition of the PI3K pathway of lung lavage cells in pneumoconiosis patients can cause the changes of cellular functions and physiological conditions via transcriptional changes, especially changes in gene expression related to immune system process, monocyte chemotaxis and leukocyte migration regulation, thus pointing to a potential key target of slowing or even pausing the pulmonary fibrosis process in pneumoconiosis patients.
Conflict of interests
The authors declare that no conflict of interests exist.
Funding
This study was supported by the National Natural Science Foundation of China (research grant of 81800071) and the Department of Science and Technology of Sichuan Province (research grant of 2019YJ0035).
References
- Farzaneh MR, Jamshidiha F, Kowsarian S (2010) Inhalational lung disease. Int J Occup Environ Med 1: 11-20.
- Cullinan P, Reid P (2013) Pneumoconiosis. Primary Care Respiratory Journal 22: 249-252.
- Luo Y, Qi XM, Pang JL, Wang J, Wang C (2021) Omics approaches for exploring pneumoconiosis: A review. Biomed Environ Sci 34: 71-82.
- Global Burden of Disease Study 2013 Collaborators (2018) Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries in 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 392: 1789-1858.
- Qi XM, Luo Y, Song MY, Liu Y, Shu T, et al. (2021) Pneumoconiosis: Current status and future prospects. Chinese Med J-Peking 134: 898-907.
- Deng CW, Zhang XX, Lin JH, Huang LF, Qu YL, et al. (2017) Association between genetic variants of transforming growth factor-beta1 and Susceptibility of Pneumoconiosis: A Meta-analysis. Chin Med J (Engl) 130: 357-364.
- Ward PA, Hunninghake GW (1998) Lung inflammation and fibrosis. Am J Respir Crit Care Med 157: 123-129.
- Keogh BA, Crystal RG (1982) Alveolitis: The key to the interstitial lung disorders. Thorax 37: 1-10.
- Borm PJ, Schins RP (2001) Genotype and phenotype in susceptibility to coal workers' pneumoconiosis. The use of cytokines in perspective. Eur Respir J Suppl 32: 127-133.
- Yucesoy B, Luster MI (2007) Genetic susceptibility in pneumoconiosis. Toxicol Lett 168: 249-254.
- Mossman BT, Churg A (1988) Mechanisms in the pathogenesis of asbestosis and silicosis. Am J Respir Crit Care Med 157: 1666-1680.
- Fujimura N (2000) Pathology and pathophysiology of pneumoconiosis. Curr Opin Pulm Med 6: 140-141.
- Kok K, Geering B, Vanhaesebroeck B (2009) Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem Sci 34: 115-127.
- Gwinn MR, Vallyathan V (2006) Respiratory burst: Role in signal transduction in alveolar macrophages. J Toxicol Environ Health B Crit Rev 1: 27-39.
- Gu W, Song J, Cao Y, Sun Q, Yao H, et al. (2013) Application of the ITS2 region for barcoding medicinal plants of selaginellaceae in pteridophyta. Plos One 8: 67818.
- Li N, Chen XW, Huo TT, Dong FQ, Deng JJ (2020) Nano-silicon dioxide affects apoptosis of alveolar macrophages via PI3K/AKt signaling pathway. Sichuan Da XueXue Bao Yi Xue Ban 51: 488-493.
- Li N, Shi F, Wang X, Yang P, Sun K, et al. ( 2021) Silica dust exposure induces pulmonary fibrosis through autophagy signaling. Environ Toxicol 36: 1269-1277.
- Liu H, Fang S, Wang W, Cheng Y, Chao J (2016) Macrophage-derived MCPIP1 mediates silica-induced pulmonary fibrosis via autophagy. Particle and fibre toxicology 13: 55.
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550.
- Wu T, Hu E, Xu S, Chen M, Guo P, et al. (2021) Cluster Profiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2: 100141.
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, et al. (2021) The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 8: 605-612.
- Weiss J, Sos ML, Seidel D, Peifer M, Zander T, et al. (2010) Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 15: 62-93.
- Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, et al. (2005) Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res 65: 3063-3071.
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, et al. (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 98: 13790-13795.
- Fernandez-Hernando C, Suarez Y (2020) ANGPTL4: A multifunctional protein involved in metabolism and vascular homeostasis. Curr Opin Hematol 27: 206-213.
- Saito M, Mitani A, Ishimori T, Miyashita N, Isago H, et al. (2020) Active mTOR in lung epithelium promotes epithelial-mesenchymal transition and enhances lung fibrosis. Am J Respir Cell Mol Biol 62: 699-708.
- Langley RJ, Mishra NC, Pena-Philippides JC, Rice BJ, Seagrave JC,et al. (2011) Fibrogenic and redox-related but not proinflammatory genes are upregulated in Lewis rat model of chronic silicosis. J Toxicol Environ Health A 74: 1261-1279.
- Giordano G, van den Brule S, Re SL, Triqueneaux P, Uwambayinema F, et al. (2010) Type I interferon signaling contributes to chronic inflammation in a murine model of silicosis. Toxicol Sci 116: 682-692.
- Pongrakhananon V, Wattanathamsan O, Takeichi M, Chetprayoon P, Chanvorachote P (2018) Loss of CAMSAP3 promotes EMT via the modification of microtubule-Akt machinery. J Cell Sci 131: 216168.
- Honma K, Chiyotani K, Kimura K (1997) Silicosis, mixed dust pneumoconiosis, and lung cancer. Am J Ind Med 32: 595-599.
- Scheffler M, Bos M, Gardizi M, Konig K, Michels S, et al. (2015) PIK3CA mutations in Non-Small Cell Lung Cancer (NSCLC): Genetic heterogeneity, prognostic impact and incidence of prior malignancies. Oncotarget 6: 1315-1326.
Citation: Luo Z, Wang M. Transcriptomic Regulation of Lung Lavage Cells from Pulmonary Fibrosis Patients under the Inhibition of PI3K. J Pulm Med Respir Res, 2021, 7: 068.
Copyright: © 2021 Zeyu Luo, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.