
Evaluation of microFLOQ™ Direct Swab for Touch DNA Recovery
*Corresponding Author(s):
Alketbi Salem KGeneral Department Of Forensic Science And Criminology, Dubai Police, Dubai, United Arab Emirates
Tel:00447774141205,
Email:alkitbe.11@hotmail.com
Abstract
This study evaluates the efficacy of the microFLOQ™ Direct Swab for touch DNA recovery from various surfaces within an office environment. Samples were collected using the swab and subjected to direct amplification with the GlobalFiler™ PCR Kit. Results revealed a success rate of 73% within three hours post-sample collection, with profiles ranging from full single to partial mixture. Despite observed artifacts, such as split and shoulder peaks, the swab demonstrated potential for efficient touch DNA recovery. However, further research is needed to assess its performance across diverse surfaces and to address challenges associated with artifact generation. Overall, the microFLOQ™ Direct Swab shows promise as a valuable tool for forensic touch DNA analysis, offering streamlined sample processing and potential advancements in forensic casework.
Keywords
Direct amplification; DNA profiling; Forensic science; Forensic investigation; GlobalFiler™ PCR Amplification Kit; MicroFLOQ™ Direct swabs; Touch DNA
Introduction
Trace or Touch DNA profiling plays a crucial role in linking individuals to criminal activities, often detected in minute quantities on various items found at crime scenes, setting it apart from other types of DNA evidence such as body fluids [1-3]. However, Touch DNA poses several challenges, including the risk of cross-contamination at crime scenes [4]. Factors such as surface types, collection methods, and environmental conditions influence the amount of Touch DNA retrieved, thereby complicating analysis [5-9]. Moreover, the extraction process and purification steps currently employed in forensic DNA casework are time-consuming and labor-intensive. Column-based purification methods can lead to DNA loss, potentially impacting the successful typing of degraded or low-copy-number samples [10-15]. Efforts to validate different collection techniques with varied methods can enhance Touch DNA recovery from a variety of surfaces [16-20].
Direct amplification in DNA profiling eliminates the need for DNA extraction and quantification, proceeding directly to PCR after sample collection. This approach not only improves recovery but also reduces the risk of errors, minimizes contamination from handling, and decreases both processing time and associated costs [1]. Recent interest has focused on developing innovative protocols for direct amplification; however, there remains limited shared knowledge regarding trace DNA collection techniques for direct PCR from casework samples [21]. One promising approach involves the use of the microFLOQ® Direct swab, a product co-developed by the French Gendarmerie Forensic Research Institute (IRCGN™) and Copan, which has shown effectiveness in collecting trace DNA for direct amplification [22-25]. The microFLOQ® Direct swabs are designed similarly to 4N6 FLOQSwabs® but are treated with a lysing agent for direct amplification, eliminating the need for DNA extraction and quantification [26]. Utilization of these swabs allows for DNA profiling from sample collection to final result in less than two hours. Additionally, due to the small dimensions of the microFLOQ® swab head, only a minimal portion of a stain is collected, resulting in significantly reduced sample consumption compared to traditional swabbing methods. Efforts to reduce sample consumption enable more evidence to be retained for retesting or post-conviction testing, if desired. Therefore, the aim of this study was to investigate the effectiveness of microFLOQ™ Direct swabs in recovering touch DNA from random surfaces of office items to replicate casework samples.
Materials And Methods
Touch samples
Biological materials were collected from a varied selection of surfaces commonly encountered in an office environment. To mimic the workflow of actual casework, these surfaces were randomly chosen, and the duration of DNA deposition was unspecified; some surfaces were routinely touched by users on a daily basis, while others had not been touched for an extended period. The sampled surfaces comprised three computer mice, three computer keyboards, two door handles, two window handles, two pens, an old handprint on a window, a leather wallet, and a cell phone (refer to Figure 1).
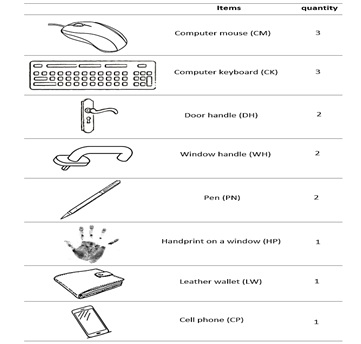 Figure 1: Office Items Utilized for Touch DNA collection to be processed for direct amplification.
Figure 1: Office Items Utilized for Touch DNA collection to be processed for direct amplification.
DNA recovery
The MicroFLOQ Direct swabs were employed to retrieve trace DNA from the surfaces outlined in Figure 1. Prior to sampling, each MicroFLOQ® Direct swab was moistened with 1μl of molecular-grade water using a pipette, following the manufacturer's instructions. Subsequently, the swab head was gently rubbed and rotated across the surface in a subsampling fashion to collect the sample.
Direct PCR amplification
Following sample collection, the tips of the MicroFLOQ® Direct swabs were detached into PCR strip tubes (0.2 ml) and subjected to amplification using the GlobalFiler™ amplification Kit (Thermo Fisher Scientific) [27], adhering to Copan's Direct DNA Analysis protocol with the microFLOQ® Collection Device [22]. This protocol involved substituting the volume of sample solution required by the kit manufacturer with molecular-grade water (15 μL), directly adding PCR master mix to the tubes (10 μL), and conducting immediate amplification on an ABI GeneAmp® 9700 PCR System (Life Technologies) for 29 cycles, following the manufacturer's recommended conditions. To evaluate potential inhibition from the lysing agent incorporated into the swab head fibers, positive control DNA (007) was amplified in the presence of a MicroFLOQ® Direct swab. Additionally, negative controls consisting solely of the MicroFLOQ® swab head were included.
DNA detection, separation, and analysis
The amplified products underwent size separation and detection on an ABI 3500 Genetic Analyzer (Life Technologies) using a mixture comprising 1 μl of PCR product, 9.6 μl of Hi-Di™ formamide, and 0.4 μl of GeneScan™ 600 LIZ® Size Standard v2.0 (Thermo Fisher Scientific). At least one microliter of allelic ladder was included per injection on the 96-well plate. Following denaturation at 95°C for 5 minutes, samples were promptly cooled on ice for 5 minutes. Electrophoresis was conducted on a 36-cm capillary array with POP-4™ polymer (Life Technologies) using standard injection parameters (1.2 kV, 24 s). Subsequently, STR data were sized and typed utilizing GeneMapper® ID-X Software Version 1.2 (Life Technologies), employing manufacturer-validated analytical thresholds.
Results And Discussion
The utilization of MicroFLOQ® swabs with the GlobalFiler™ amplification Kit for direct PCR, aimed at recovering Touch DNA from assorted items within an office setting, yielded a success rate of 73% within a three-hour timeframe post-sample collection. Out of the 15 samples collected, 11 STR profiles yielded positive DNA results suitable for database search. These profiles comprised full single, partial single, full mixture, and partial mixture DNA profiles (refer to Figures 2&3). However, the majority of profiles exhibited artifacts such as split and shoulder peaks. As per the criteria set by the Biology and DNA Section lab within the General Department of Forensic Science and Criminology, positive DNA results for forensic trace profiles necessitate homozygous or heterozygous alleles, or a combination thereof, in a minimum of nine loci. The negative control employed in the DNA profiling process was confirmed to be free of DNA. Additionally, amplification of Positive control DNA (007) in the presence of a MicroFLOQ® swab head resulted in complete STR profiles, demonstrating expected performance. This outcome suggests that the chemical treatment of MicroFLOQ® swab heads with a lysing agent by the manufacturer does not impede or inhibit successful DNA amplification.
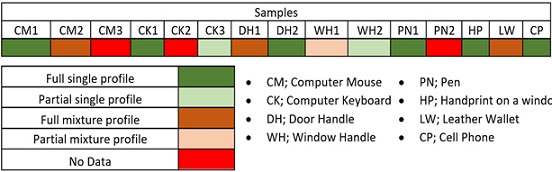 Figure 2: Touch DNA profiles (n= 15) obtained from office items using the MicroFLOQ Direct swab and directly amplified through the GlobalFiler™ PCR Kit. The samples yielded 6 complete single DNA profiles, 2 partial single DNA profiles, 3 complete mixture DNA profiles, 1 partial DNA profile, and 3 profiles with no data.
Figure 2: Touch DNA profiles (n= 15) obtained from office items using the MicroFLOQ Direct swab and directly amplified through the GlobalFiler™ PCR Kit. The samples yielded 6 complete single DNA profiles, 2 partial single DNA profiles, 3 complete mixture DNA profiles, 1 partial DNA profile, and 3 profiles with no data.
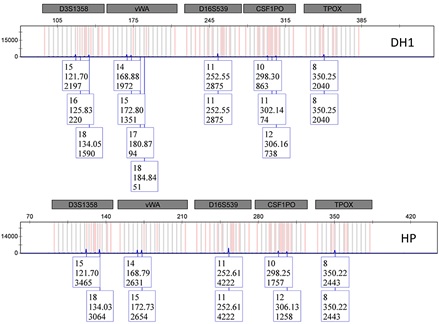 Figure 3: Electropherograms of samples gathered from a door handle (DH1), resulting in a complete mixture DNA profile, and from a visible handprint on the window (HP), yielding a complete single DNA profile. These profiles highlight variations in peak height across five autosomal STR loci (D3S1358, vWA, D16S539, CSF1PO, and TPOX).
Figure 3: Electropherograms of samples gathered from a door handle (DH1), resulting in a complete mixture DNA profile, and from a visible handprint on the window (HP), yielding a complete single DNA profile. These profiles highlight variations in peak height across five autosomal STR loci (D3S1358, vWA, D16S539, CSF1PO, and TPOX).
Previous research by Templeton et al. [28] compared the performance of foam swabs, cotton swabs, and nylon FLOQSwabs® in collecting trace DNA from fingerprints, with the latter demonstrating the highest DNA yield. This study was expanded to include the use of FLOQSwabs® for touch DNA recovery across various substrates, comparing direct amplification to the standard extraction workflow. Findings from these controlled experiments underscore the potential of the FLOQSwab® system and direct amplification in enhancing signal retrieval from low-level touch samples, while conserving resources and mitigating contamination risks [29].
Moreover, the adoption of direct detection facilitated a rapid DNA profiling process, albeit accompanied by the observation of artifacts such as split and shoulder peaks, consistent with prior reports [23-24]. Distinguishing true alleles within DNA profiles can be straightforward when originating from a single source, yet becomes complex in mixed DNA profiles, particularly in the absence of reference samples.
The results obtained from this limited set of touch samples suggest that the miniaturized microFLOQ® swab head can retrieve sufficient cellular material to generate complete DNA profiles from diverse surfaces. However, the quantity of material present and the nature of the surface influence the likelihood of success, as anticipated for evidence collection via swabbing.
Conclusion
In conclusion, the findings of this study provide valuable insights into the effectiveness of the MicroFLOQ™ Direct Swab for Touch DNA recovery. While the results demonstrate promising success rates within a controlled office environment, further research is necessary to comprehensively evaluate its efficiency across a broader spectrum of surfaces and scenarios. Additionally, the observed artifacts, such as split and shoulder peaks, underscore the need for future work to develop strategies to mitigate these challenges associated with direct amplification. By addressing these areas of improvement, the MicroFLOQ™ Direct Swab has the potential to become an invaluable tool in forensic investigations, offering enhanced efficiency and reliability in Touch DNA recovery.
Conflict of interest
None.
Acknowledgements
The authors would like to express their gratitude to the General Department of Forensic Science and Criminology in Dubai Police for approving this study. Ethical approval was granted by the University of Central Lancashire's Research Ethics Committee (ref. no. STEMH 912), for which we are sincerely thankful.
References
- Alketbi SK (2018) The affecting factors of Touch DNA. J of Forensic Res 9: 424.
- Alketbi SK (2023) Analysis of Touch DNA. Doctoral thesis, University of Central Lancashire.
- Alketbi, SK (2023) The role of DNA in forensic science: A comprehensive review. Int J of Sci and Res Arch 9: 814-829.
- Alketbi SK (2023) Maintaining the chain of custody: Anti-contamination measures for trace DNA evidence. International J Sci Res Arch 8: 457-461.
- Alketbi SK, Goodwin W (2019) The effect of surface type, collection, and extraction methods on Touch DNA. Forensic Science International. Genetics Supplement Series 7: 704-706.
- Alketbi SK, Goodwin W (2019) The effect of time and environmental conditions on Touch DNA. Forensic Science International. Genetics Supplement Series 7: 701-703.
- Alketbi SK, Goodwin W (2019) The effect of sandy surfaces on Touch DNA. Journal of Forensic Legal & Investigative Sciences 5: 034.
- Alketbi SK (2020) Collection of Touch DNA from rotten banana skin. International Journal of Forensic Sciences 5: 000204.
- Alketbi SK (2023) An Evaluation of the Performance of Two Quantification Methods for Trace DNA Casework Samples. J Forensic Sci Criminal Invest 16: 555950.
- Barta JL, Monroe C, Teisberg JE, Winters M, Kemp BM, et al. (2014) One of the key characteristics of ancient DNA, low copy number, may be a product of its extraction. J of archaeol sci 46: 281-289.
- Mumy KL, Findlay RH (2004) Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative-competitive PCR. J microbiol methods 57: 259-268.
- Dabney J, Knapp M, Gansauge MT, Weihmann A, Nickel B, et al. (2013) Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proceedings of the National Academy of Sciences - PNAS 110: 15758-15763.
- Kemp BM, Winters M, Monroe C, Barta JL (2014) How Much DNA is Lost? Measuring DNA Loss of Short-Tandem-Repeat Length Fragments Targeted by the PowerPlex 16® System Using the Qiagen MinElute Purification Kit. Human biology 86: 313-329.
- Doran AE, Foran, DR (2014) Assessment and mitigation of DNA loss utilizing centrifugal filtration devices. Forensic science international: Genetics 13: 187-190.
- Garvin AM, Fritsch A (2013) Purifying and Concentrating Genomic DNA from Mock Forensic Samples Using Millipore Amicon Filters. Journal of forensic sciences 58: S173-S175.
- Alketbi SK, Goodwin W (2021) Touch DNA collection techniques for non-porous surfaces using cotton and nylon swabs. Biomedical Journal of Scientific & Technical Research 36: 28608-28612.
- Alketbi SK (2022) The impact of collection method on Touch DNA collected from fabric. Journal of Forensic Sciences and Criminal Investigation 15: 555922.
- Alketbi SK, Goodwin W (2022) The impact of area size and fabric type on Touch DNA collected from fabric. Journal of Forensic Sciences and Criminal Investigation 16: 555926.
- Alketbi SK, Goodwin W (2022) The impact of deposition area and time on Touch DNA collected from fabric. Forensic Science International, Genetics Supplement Series 8: 45-47.
- Alketbi SK (2023) Collection techniques of touch DNA deposited on human skin following a strangulation scenario. International Journal of Legal Medicine 137: 1347-1352 .
- Cavanaugh SE, Bathrick AS (2018) Direct PCR amplification of forensic touch and other challenging DNA samples: A review. Forensic science international: Genetics 32: 40-49.
- Ambers A, Wiley R, Novroski N, Budowle B (2018) Direct PCR amplification of DNA from human bloodstains, saliva, and touch samples collected with microFLOQ® swabs. Forensic science international: Genetics 32: 80-87.
- Alketbi SK, Goodwin W (2023) Collection Methods for Touch DNA Direct Amplification. Journal of Forensic, Legal and Investigative Sciences 9: 072.
- Alketbi SK (2022) An innovative solution to collect Touch DNA for direct amplification. Journal of Forensic Sciences and Criminal Investigation 16: 555928.
- Alketbi SK, Alsoofi S (2022) Dual recovery of DNA and fingerprints using Minitapes. Journal of Forensic Sciences and Criminal Investigation 16: 555929.
- microFLOQ® Direct product brochure, https://www.copangroup.com/product-ranges/microfloq/
- GlobalFiler™ PCR amplification kit user guide. (2016) Revision E.
- Templeton J, Blackie R, Viviana P, Oliva H, Taylor D (2013) Genetic profiling from challenging samples: Direct PCR of touch DNA. Forensic science international. Genetics supplement series 4: e224-e225.
- Templeton JE, Taylor D, Handt O, Skuza P, Linacre A (2015) Direct PCR Improves the Recovery of DNA from Various Substrates. Journal of forensic sciences 60: 1558-1562.
Citation: Alketbi SK, Goodwin W (2024) Evaluation of microFLOQ™ Direct Swab for Touch DNA Recovery. Forensic Leg Investig Sci 10: 093.
Copyright: © 2024 Alketbi Salem K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

