
Genetic Incompatibility between Father and Child Due To Maternal Uniparental Isodisomy at Locus D13S317 in A Paternity Testing: A Case Report
*Corresponding Author(s):
Anshu PriyaAssistant Professor, Forensic Finger Printing Lab, Centre For Advanced Research, KGMU Lucknow, India
Email:anshupriya@kgmcindia.edu
Abstract
Uniparental Disomy (UPD) is a rare type of chromosomal aberration that has sometimes been detected in paternity testing. In the present scenario, one such case was observed at the forensic science laboratory, in Ranchi. Here we examined a 3-person family [deceased (claimed son), father, and mother] by using 15 autosomal short tandem repeat markers, along with gender locus using AmpFlSTR®Identifiler® PCR amplification kit, further to enhance the accuracy and reliability of parentage testing, Y STR profiling was performed which provides a powerful method to detect a UPD. We used an AmpFlSTR®YfilerTM PCR amplification kit to conduct Y STR DNA analysis because the child was male. In the present study, we observed that marker D13S317 on chromosome 13 was not transmitted in a Mendelian style between the father and the claimed son. However, the maternal allele was present at all the corresponding loci. The composition of the alleles of this locus in the father, claimed child and mother was 10/10, 11/11, and 11/12 respectively. Interestingly Chromosome13 in the claimed son showed a complete maternal UPD. To the best of our knowledge, this is a rare case of maternally transmitted isodisomy reported for the first time for this locus in forensic DNA analysis. The test results made it impossible to rule out the deceased person's identification, although they may be an instance of uniparental maternal disomy.
Keywords
Paternity testing; Short tandem repeat profiling; Uniparental isodisomy
Introduction
DNA profiling of Short Tandem Repeat (STR) is widely used in forensic sciences, including paternity testing and kinship analysis [1]. In forensic laboratories, DNA testing in cases of disputed paternity is a standard procedure. The child inherits half of its DNA from each of its biological parents, which is the fundamental premise of genetic analysis in cases of disputed paternity. The absence of shared alleles in the analyzed loci between the child and the claimed father may point to the examined man's exclusion from paternity. Uniparental Disomy (UPD) is referred to as both homologous chromosomes inherited from only one parent. It can originate from the father (UPD pat) or the mother (UPD mat). UPD can be further classified as isodisomy and heterodisomy. Isodisomy is defined as two identical copies of one allele of a single parent and heterodisomy is defined as two different alleles of the contributing parent. An individual inherits one copy of a Short Tandem Repeat (STR) from each parent, which may or may not have similar repeat sizes.
The number of repeats in STR markers can be highly variable among individuals, but stably inherited from parents to children and follows simple Mendelian inheritance, which makes these STRs effective for human identification purposes [2]. However, genomic variations in the regions of STR loci can result in an allelic mismatch in the questioned child, which may complicate the forensic inference in the case of paternity testing. DNA testing is a common procedure in genetic laboratories when there is a paternity question. The test's objective is to show the father, mother, and the subject child's studied genetic markers' similarities and differences. It may be possible to rule out biological parenthood if there are differences in the studied loci between the putative child and the father. Genetic mutations, such as chromosome abnormalities, are another cause of these variations. DNA fingerprinting is now frequently used to resolve paternity disputes. In many paternity cases, the father denies fatherhood to the child, either in an illicit relationship or during pregnancy as a result of rape, according to forensic research.
In this scenario, the present case was taken from the person who was murdered during the Naxalite encounter and had to establish the identity of the person. In such legal cases, the court typically demands paternity testing to be performed to confirm the truth, although in DNA profiling, the mutation is one aspect that is occasionally found and makes problems difficult to resolve. Y DNA profiling is a helpful approach to determine the progeny and paternity when there is a male child and a mismatch is noticed since the Y DNA is only passed from man to male and is the same for a child, father, uncle, and brother who are from the same progeny.
- Case presentation
We present a case of an unknown deceased person (a claimed son) who was murdered during the Naxalite encounter, in Jharkhand during the investigation some blood-stained earth along with stone pieces were collected and sent to the forensic science laboratory for examination to establish the identity of the person. Paternity testing was obtained for legal reasons, samples from the father (source: blood-positive gauze piece cutting) and an unknown deceased person (source: blood-positive stone piece swab cuttings) were analyzed using Identifiler plus kit with standard laboratory practices. A total of 15 polymorphic loci were genotyped. Initial results appeared to exclude the father from paternity due to genetic inconsistencies at loci D13S317. However, we performed additional testing, and the Y-str profiling of the deceased person and father had been done to rule out parenthood. But we could not get the complete profile of the deceased person. However, we demanded a reference sample of the mother to establish his identity and it was found that the deceased was homozygous for maternal alleles on chromosome 13.
Materials And Methods
- DNA extraction
Samples from the father (source: blood-positive gauze piece cutting) and unknown deceased person (source: blood-positive stone piece swab cuttings) and mother (source: blood-positive gauze piece cutting) were all soaked in 400 ul of extraction buffer (10 mM Tris-Cl, pH 8.0, 0.1M EDTA, pH 8.0, 20 g/ml RNase A, 0.5% SDS) for an hour at 37°C After that, Proteinase K was added, and the mixture was continuously swirled for a further three hours at 56°C. A similar volume of Tris-equilibrated phenol (pH 8.0) was added to the solution, which had been warmed to room temperature. The tube was then turned end over end for five minutes to mix the ingredients. The tubes were then centrifuged at 5000 X g for 10 min at room temperature. The supernatant was then transferred to another fresh tube and DNA was precipitated by adding 2X vol of absolute ethanol at -20°C for 30 min. The tubes were then centrifuged at 15, 000 X g for 15 min. The obtained pellet was washed with 70% ethanol and dried at room temperature. The DNA was resuspended in the desired volume of TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8.0) and stored at 4°C.
- DNA quantification
Quantifiler® Human DNA Quantification kit (Life Technologies Inc.) containing DNA standard solution (200 ng/μl), Quantifiler Human Primer mix, and Quantifiler PCR Reaction Mix was used to quantify the human DNA with real-time PCR (RT-PCR). To create a 25µl PCR reaction system, human primer mix (10.5 µl/sample) and PCR reaction mix (12.5 µl/sample) were mixed and then dispensed into reaction wells (23 µl each) [3]. Next, 2 µl of a sample or standard DNA of known concentration was added to each well. The amount of DNA was calculated by the real-time PCR machine (Applied Biosystems).
- Identifiler plus® PCR reaction
A multiplex PCR reaction for the simultaneous amplification of 15 autosomal STR loci and a gender locus was conducted using the AmpFISTR Identifiler Plus® kit [4]. (Table 1). Amplification was done with 25 µl of PCR amplification mixture (10.5 µl of PCR reaction mix, 5.5 µl of Identifiler Plus® Primer Set, 9.0 µl of nuclease-free water, and 1 µl of DNA template), under the following conditions: initial denaturation at 95° C for 10 min; followed by 29 cycles of denaturation at 94° C for 1 min; annealing at 59° C for 1 min and extension at 72° C for 1 min, and a final extension step at 60° C for 60 min.
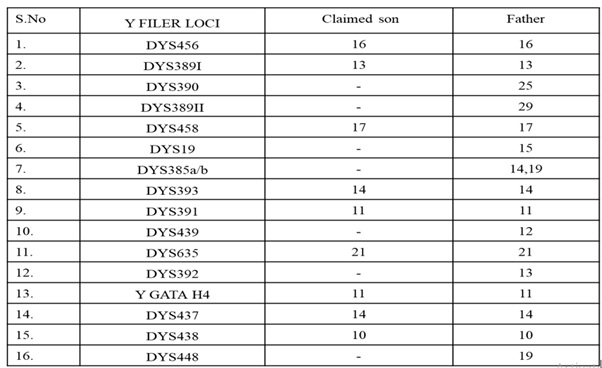 Table 1: Examination of 16 Y-STR Markers.
Table 1: Examination of 16 Y-STR Markers.
Capillary Electrophoresis
The PCR product was then evaluated in electrophoresis equipment that contained 1µl of PCR product or the AmpFlSTR® Identifiler Plus® allelic ladder, 0.3 µl of GeneScanTM 500 LIZ® Size Standard dye, and 8.7 µl of Hi-DiTM formamide. A 36 cm 4-capillary array (Applied Biosystems) was used for the capillary electrophoresis, which was carried out using a POP-4 polymer and an ABI-3130 Genetic Analyzer. Gene Mapper ID software version 3.2 was used to analyse data in relation to the Gene ScanTM 500 LIZ® Size Standard.
- Y-STR amplification from blood stain deceased (claimed son) and father’s references samples
AmpFlSTR®YfilerTM PCR amplification kit was used to genotype the father’s reference sample and the claimed son. The reactions were performed as described above, except the number of PCR cycles that were reduced to 30 in all instances. Moreover, a total of 0.5-1.0 ng of DNA was used per PCR reaction.
Results
A commercially available STR kit's 15 standard STR markers and one gender locus amelogenin is being used for the paternity test in all the forensic laboratory followed by capillary electrophoresis. The allelic distribution (genotypes) obtained after the data collection analysed by software and shown in (Figure 1) It displays all the investigated loci across all the exhibits by the Gene mapper software (Applied Biosystems) and (Table 2) displays a comparative chart of the allele distribution (genotype) of the various autosomal DNA loci. Following the initial study (15 DNA STRs made using the AmpFlSTR1 Identifiler), a discrepancy at locus D13S317 (located on chromosome 13) was found and shown in (Figure 2).
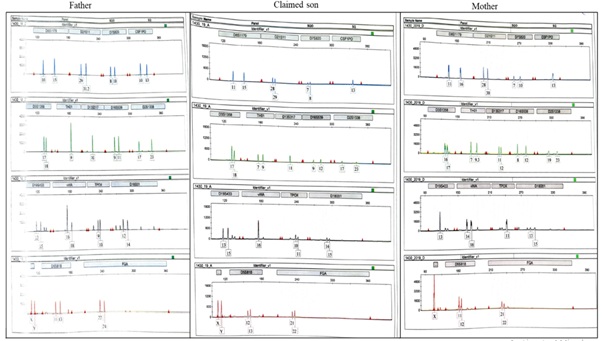 Figure 1: Electropherogram of genotypes of the father, the claimed son, and the mother at 15 different loci of the autosomal DNA.
Figure 1: Electropherogram of genotypes of the father, the claimed son, and the mother at 15 different loci of the autosomal DNA.
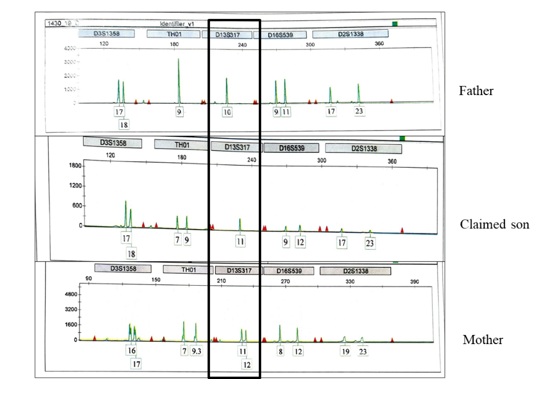 Figure 2: Electropherogram of genotypes of the father, the claimed son, and the mother at text locus D13S317.
Figure 2: Electropherogram of genotypes of the father, the claimed son, and the mother at text locus D13S317.
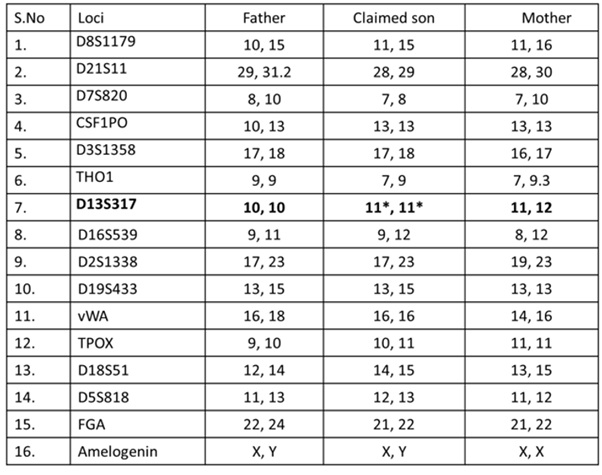 Table 2: Comparative chart of allele distribution (genotype) of different loci of the autosomal DNA tested.
Table 2: Comparative chart of allele distribution (genotype) of different loci of the autosomal DNA tested.
*Mutated allele shows the mismatch with the father
The Additional Y STRs profiling using the Y Filer kit were analyzed for the father and the claimed son because the Y chromosome is only found in males, which are only passed down by the father, making the Y chromosome in any paternal line practically identical because one exclusion was not considered sufficient to declare non-paternity in such type of case analysis.
However, this step did not reveal a complete DNA profile, which is shown in (Figure 3) and the Comparative chart of allele distribution (genotype) of different Y STR loci is shown in (Table 1). However, this study revealed that the alleged son had the maternal allele at all the loci except locus D13S317, as shown in (Table 2). The father, the alleged child, and the mother each had 10/10, 11/11, and 11/12 alleles at the D13S317 locus. Intriguingly, the claimed son's Chromosome 13 displayed a full maternal UPD. The results noted above and the claimant's son and mother sharing the same chromosome 13 genotype that supported maternal Uniparental Disomy (UPD13).
 Figure 3: Electropherogram of genotypes of the claimed son & father the 17 Y-STR Markers.
Figure 3: Electropherogram of genotypes of the claimed son & father the 17 Y-STR Markers.
Discussion
In the paternity case presented here, we observed a genetic inconsistency at chromosome 13. It is highly unusual to perform parentage testing in the forensic laboratory for such novel cases. We aimed to pursue further testing because of the genetic inconsistencies between the claimed son and the father at locus D13S317 which was observed during the initial autosomal testing approach. One inconsistency could have represented actual exclusions or genetic variants, so this necessitated further testing. Nevertheless, we were convinced that the claimed son was the biological son. Using the PCR Amplification technique, the nuclear DNA profile of the father, claimed son, and mother was generated. Interestingly it was observed that paternal alleles were present in claimed son at14 STR loci except for one locus ‘D13S317’ among 15 different genetic loci which might be caused due to mutation. We cannot exclude the child of the father as a true child if he eliminates two genetic loci according to two exclusion standards in parentage testing. However, the mother's STR loci were all matched. In order to rule out paternity in this situation, we also used Y DNA profiling to establish whether the child's progeny and the father were related. Through the use of analysis to detect mutations in the nuclear DNA profile, paternal lineage can be determined. Additionally, the typing results for chromosome 13 known as Uniparental Disomy 13 were remarkably consistent with a solely maternal origin (UPD13).
A rare disorder known as uniparental disomy develops when a child receives no copy from one parent and two copies of one or more chromosomes from one parent [5]. We don't know how frequently this occurs, and specifically, the particular form of UDP 13, occurs. UPD may result from a variety of genetic factors, including the loss of a chromosome from an initial trisomy zygote, the duplication of a chromosome from a monosomy zygote, and the fertilization of a gamete with two copies of a chromosome by a gamete without a copy of the homologous chromosome, a process known as gamete complementation and might be also because of post-fertilization error like the non-disjunction of sister chromatids during mitosis after fertilization [6]. For the first time, a maternal uniparental isodisomy at chromosome 13 is shown in this study. When both chromosome 13 were homologously recombined during maternal meiosis, the observed homoallelic pattern of the child's second chromosome 13 may be explained. The next non-disjunction occurrence during the first meiotic division most likely caused an egg to be disomic for chromosome 13. So, the formation of the present UPD13 could have been caused by either trisomy rescue or gamete complementation. Robinson's research [7] estimates the incidence of UPD to be about 0.029% (1 in 3500). The incidences of UPD for various chromosomes appear to vary significantly.
Conclusion
The study conclusively proved that the mismatch of the alleles of D13S317 locus in the child with the father is might be due to the UPD & the claimed child could be the biological son of the claimant parents. Our work emphasize that pseudo-exclusion might occur if the inconsistent loci were all on the same chromosome. In this situation, investigators must consider the possibility of UPD specially in case of male child. Our study also suggests that additional alternative method such as Chromosomal Microarray (CMA) could be used to validate our study. Next generation sequencing, with evenly distributed loci on that chromosome, could be an effective method to detect UPD, helping investigators to draw conclusions in parentage tests.
Declaration of Competing Interest
The authors have no conflict of interest
References
- Henke L, Henke J (2005) Which short tandem repeat polymorphisms are required for identification? Lessons from complicated kinship cases. Croat Med J 46: 593-597.
- Liehr T (2010) Cytogenetic contribution to uniparental disomy. Mol Cytogenet 3: 8.
- Green RL, Roinestad IC, Boland C, Hennessy LK (2005) Developmental validation of the quantifiler real-time PCR kits for the quantification of human nuclear DNA samples. J Forensic Sci 50: 809-825.
- Qiu-Ling L, De-Jian L, Li Q, Ye-Fei C, Shen M, et al. (2012) Development of multiplex PCR system with 15 X-STR loci and genetic analysis in three nationality populations from China. Electrophoresis 33: 1299-1305.
- Engel E (1980) A new genetic concept: Uniparental disomy and its potential effect, isodisomy. Am J Med Genet 6: 132-143.
- Kotzot DG (2005) Utermann, Uniparental Disomy (UPD) other than 15: Phenotypes and bibliography updated Am. J Med Genet 136A: 287-305.
- Robinson WP (2000) Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays 22: 452-455.
Citation: Priya A, Kumar A, Bara N, Soren AN (2023) Genetic Incompatibility between Father and Child Due To Maternal Uniparental Isodisomy at Locus D13S317 in A Paternity Testing: A Case Report. J Forensic Leg Investig Sci 9: 074.
Copyright: © 2023 Anshu Priya, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

