
Proteomics Methods in Meat Research: A Review of Applications for Species Authentication, Quality Evaluation, and Adulteration Detection
*Corresponding Author(s):
Zubayed AhamedTasmanian Institute Of Agriculture, University Of Tasmania, Australia
Email:zubayed.nft.just@gmail.com
Abstract
Proteomics has emerged as a transformative approach for molecular characterization of meat quality, authenticity, and safety in modern food systems. By enabling comprehensive profiling of the muscle proteome, it offers exceptional sensitivity and specificity in detecting protein biomarkers linked to key quality traits, species authentication, and adulteration detection. Advances in proteomic technologies, including two-dimensional gel electrophoresis, Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS), and data-independent acquisition, have expanded the ability to identify and quantify proteins, post-translational modifications, and degradation products relevant to meat science. Integration with advanced bioinformatics tools enhances interpretation of complex datasets, supporting the development of robust quality markers and predictive models. Despite these advances, challenges remain, such as the complexity of meat matrices, limited protein databases, and the need for standardized analytical protocols. Continued innovation in instrumentation, database enrichment, and methodological harmonization will be vital for realizing the full potential of proteomics in meat quality assurance and authentication.
Keywords
Meat adulteration; Meat quality; Proteomics; Species authentication; Spectrometry
Introduction
Meat remains a cornerstone of human nutrition, providing not only high-quality protein but also essential vitamins, minerals, and amino acids necessary for optimal health [1]. Recent research has significantly contributed to the understanding of various aspects of meat quality and animal production, including the use of probiotics [2], freezing impacts on organ meat [3], and meat adulteration detection using NIRS and chemometric analysis [4]. Furthermore, studies have explored innovative technologies like machine learning and AI for improving livestock management and meat safety evaluation [5,6]. Additional reviews have addressed heat stress [7], poultry processing [8], and cutting-edge technology meat quality and meat preservation techniques [9,10]. These collective efforts underscore the growing reliance on interdisciplinary approaches for enhancing meat quality, food safety, and animal productivity. The modern meat industry is tasked with upholding rigorous standards for quality, safety, and traceability, as these attributes are central to consumer trust and regulatory compliance. Among these, meat quality stands out as the primary determinant of consumer satisfaction, especially in regions such as Europe, where expectations for authenticity and product excellence are particularly high [11]. However, achieving consistent meat quality is inherently complex. Variability in animal genetics, feed composition, husbandry practices, processing technologies, and storage conditions can all contribute to significant fluctuations in key sensory attributes-texture, flavor, color, and shelf life. Such inconsistencies not only shape consumer perceptions but also pose risks to industry reputation and market stability.
Incidents like the horse meat scandal, where horse meat was fraudulently sold as beef across several European countries, have exposed vulnerabilities in meat supply chains and intensified public concern over food fraud, authenticity, and traceability [12]. In response, the industry has increasingly turned to advanced analytical technologies to safeguard product integrity. Among these, proteomics has emerged as a transformative approach for meat quality assessment, species authentication, and adulteration detection [13,14]. By enabling comprehensive profiling of the meat proteome, proteomics provides molecular-level insights into the factors that define quality traits and track biochemical changes throughout processing, storage, and cooking [15]. Unlike DNA-based methods, proteomic analyses capture the dynamic postmortem modifications and processing-induced changes in proteins, making them especially valuable for evaluating processed meat products [16].
Recent studies have established direct links between proteomic signatures and the generation of flavor compounds in meat [17]. Flavor, a critical driver of consumer acceptance, arises from intricate interactions among volatile and non-volatile molecules produced during heating, storage, and processing [18]. These interactions are modulated by proteolytic enzymes and amino acid catabolism, both of which are reflected in the meat’s proteomic profile. Key biochemical pathways, including the Maillard reaction, Strecker degradation, and lipid oxidation, further shape the sensory landscape of meat products [19]. Beyond flavor, proteomics enables the evaluation of other vital quality indicators such as water-holding capacity, oxidative stability, tenderness, and color [20]. Biomarkers like calpains, cathepsins, and myofibrillar protein fragments are closely associated with postmortem tenderization and aging, while the detection of spoilage-related proteins and oxidative stress markers supports more accurate shelf-life prediction and food safety assurance [21,22].
The extraction and analysis of meat proteins rely on sophisticated methodologies, including two-Dimensional Electrophoresis (2-DE), Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS), and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) [16]. LC-MS/MS, in particular, offers high sensitivity and the ability to detect low-abundance proteins in complex matrices [23]. These workflows typically involve protein extraction, enzymatic digestion, and peptide separation, followed by advanced data analysis using multivariate statistical tools such as Principal Component Analysis (PCA), hierarchical clustering, and pathway enrichment analysis [24]. Such computational approaches not only facilitate the discrimination of meat types and treatments but also provide mechanistic insights into the underlying biological processes, positioning proteomics as a powerful complement to other omics technologies like volatilomics [25].
Despite its promise, the application of proteomics in meat science is not without challenges. The complexity of meat matrices, diversity of post-translational modifications, and susceptibility of proteins to degradation during processing all present analytical hurdles. However, ongoing advancements in mass spectrometry instrumentation and bioinformatics are steadily overcoming these obstacles, enabling more robust, high-throughput, and accurate analyses [16]. This review synthesizes recent scientific progress and practical applications of proteomics in meat research, with a focus on quality assessment, species authentication, and adulteration detection. It also addresses current limitations and outlines future directions for integrating proteomics into routine industry practice, ultimately aiming to enhance quality control, foster consumer confidence, and support regulatory compliance in the global meat sector.
Current Research in Meat Quality Assessment and Its Limitations
Recent advances in meat quality assessment have largely centered on evaluating sensory properties, traditional quality factors, and the physical and chemical characteristics of meat. Conventional approaches for determining meat freshness include microbiological analysis, Total Volatile Basic Nitrogen (TVB-N) measurements, and gas chromatography [26]. While these methods are widely used, they present several inherent limitations.
Subjectivity in Sensory Evaluation
Sensory quality attributes, such as flavor, texture, and aroma, are typically assessed by trained panels or consumers. However, these evaluations are inherently subjective, influenced by individual perception, experience, and physiological differences, which can lead to inconsistent and non-reproducible results [27].
Microbiological and Chemical Analyses
Standard microbiological tests are effective for detecting mesophilic bacteria but often fail to identify psychrotrophic bacteria that can proliferate during cold storage, thereby limiting their reliability for comprehensive spoilage detection [28]. TVB-N measurements and similar chemical assays provide useful indicators of spoilage but may not capture early or subtle changes in meat quality.
Instrumental Techniques
Gas chromatography is highly accurate for identifying volatile spoilage markers, yet it is cost-prohibitive, labor-intensive, and requires specialized technical expertise. This restricts its application in routine, high-throughput, or online quality control settings. Furthermore, while the human olfactory system can detect a wide range of odors through approximately 400 types of receptors, sensory assessments remain subjective and are affected by individual bias and sensitivity [29]. Importantly, the human nose cannot detect odorless but potentially harmful compounds, limiting its effectiveness as a consistent tool for spoilage or safety classification [27].
Emergence of Proteomics in Meat Quality Assessment
To address the persistent challenges in meat quality evaluation, the field of meat science is increasingly adopting proteomics-a powerful and comprehensive approach that investigates the entire set of proteins expressed in a biological system under specific physiological or environmental conditions [30]. Traditional meat quality assessment methods, such as physicochemical measurements, sensory evaluation, and microbial tests, often lack the sensitivity and consistency required for high-precision analysis. In contrast, proteomics provides an advanced molecular-level understanding of biological processes that underlie meat quality attributes. By studying proteins, which serve as the functional molecules of cells, proteomics allows researchers to uncover the biochemical mechanisms that influence tenderness, color, water-holding capacity, flavor, and overall consumer acceptability. Furthermore, the integration of proteomics into meat science represents a paradigm shift, offering not only analytical precision but also novel opportunities for predictive modeling, authentication, and quality assurance across the entire meat production and supply chain.
Objectivity and Reproducibility
Unlike conventional sensory or microbial methods, which can sometimes be delayed, biased, or influenced by environmental variability, proteomics provides objective, reproducible, and molecular-level data for evaluating meat quality [31]. This objectivity stems from the quantifiable and highly specific nature of protein expression and modification patterns. Proteomic techniques minimize human subjectivity and enable standardization across laboratories and production facilities, which is essential for generating reliable and comparable results worldwide. The reproducibility of proteomic data also ensures that consistent conclusions can be drawn from independent experiments, making it particularly valuable for establishing universal markers of meat quality.
Comprehensive Protein Profiling
Proteomic analyses allow for the identification and quantification of a wide range of proteins, along with insights into their post-translational modifications, degradation patterns, and protein–protein interactions [32]. These detailed molecular fingerprints reflect the dynamic biochemical events that occur during various stages of meat production-slaughter, storage, aging, and cooking. For instance, proteolysis and protein oxidation during post-mortem aging directly affect tenderness and juiciness, while structural protein degradation influences water-holding capacity and drip loss. By mapping these molecular changes, proteomics provides a holistic view of meat quality determinants, enabling scientists to link specific protein profiles with desirable or undesirable traits. Such insights can help optimize meat processing techniques, improve shelf-life prediction, and develop targeted interventions to maintain product quality.
Early Detection and Predictive Quality Control
One of the defining advantages of proteomics is its ability to detect subtle biochemical changes in meat at an early stage, long before spoilage or quality deterioration becomes evident through sensory or chemical analyses [33]. Specific protein degradation products, oxidative markers, or microbial contamination signatures can be identified as early biomarkers that indicate the onset of biochemical instability. These biomarkers allow producers to implement predictive quality control strategies, ensuring that corrective actions such as improved packaging, antioxidant supplementation, or modified storage conditions are applied before significant losses occur. Moreover, proteomic data can be incorporated into predictive models, providing processors and retailers with actionable insights to optimize logistics and extend product shelf life. This proactive approach represents a major improvement over reactive quality assessments that only detect issues after consumer complaints or visible spoilage.
Authentication and Traceability
Proteomic profiling has also emerged as a highly reliable tool for authentication and traceability in the global meat industry. It can accurately distinguish between species and breeds by identifying species-specific or breed-specific protein markers [30]. This capability is particularly valuable for detecting fraudulent substitutions, such as the illegal inclusion of horse meat in beef products, which can have significant implications for consumer trust, religious dietary restrictions, and regulatory enforcement. Beyond fraud detection, proteomics ensures transparency across the supply chain by confirming the declared origin and production system of meat products. In regions with a history of food scandals, the adoption of proteomic authentication methods helps rebuild consumer confidence while enabling regulatory bodies to enforce strict quality standards. Ultimately, proteomics strengthens the link between product labeling, consumer expectations, and actual product composition, ensuring both safety and credibility in meat markets.
Research And Innovation Enabled By Proteomics
Beyond routine quality assessment, proteomics is instrumental in advancing research and innovation. It allows scientists to unravel how genetic, environmental, and dietary factors influence muscle protein composition, supporting the development of improved breeding strategies, optimized processing conditions, and targeted interventions using natural additives or antioxidants. The increasing accessibility of high-throughput techniques such as LC-MS/MS, 2D-GE, and shotgun proteomics is accelerating the integration of proteomic tools into meat science, making them indispensable for ensuring quality, safety, and consumer confidence in the global meat industry.
Proteome Technologies in Meat Science
Advancements in proteomics technologies have fundamentally transformed the study of protein expression, function, and interaction within biological systems, providing unprecedented insights into the molecular determinants of meat quality (Table 1). These high-throughput platforms enable the comprehensive identification and quantification of proteins in complex matrices such as muscle tissue, which is essential for elucidating the biochemical basis of key quality traits.
|
Technologies |
Principle |
Significance |
References |
|
4D label-free quantitative proteomics |
Advanced technology integrating ion mobility spectrometry with traditional mass spectrometry to analyze protein profiles and metabolic pathways related to meat quality. |
Detects subtle protein/metabolite changes during storage, processing, or spoilage. |
Xu et al. [34] |
|
Label-free quantitative proteomics technology |
Protein abundance is measured by comparing peptide signal intensities without chemical or isotopic labeling. |
Applied to study protein degradation, oxidative modifications, and muscle-to-meat conversion. |
Lin et al. [35] |
|
Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS) |
Liquid chromatography separates peptides; tandem MS identifies and quantifies them based on fragmentation patterns. |
Used in the meat industry for many purposes, particularly for quality assessments |
Zhang et al. [36] |
|
4D-DIA technology |
Data-Independent Acquisition combined with 4D separation allows comprehensive and reproducible proteome coverage. |
The capability to find out the protein biomarkers from different meat quality traits |
Sun et al. [37] |
|
SDS-PAGE |
Separates proteins based on molecular weight using sodium dodecyl sulfate–polyacrylamide gel electrophoresis. |
Extraction of protein, such as sarcoplasmic and myofibrillar proteins |
Govindaiah et al. [38] |
|
Tandem Mass Tag (TMT)-10plex labelling-based proteomics. |
Uses isobaric chemical tags to label peptides, allowing multiplexed quantification of up to 10 samples in one run. |
Evaluation of quality attributes (with different color stability, tenderness, and Water holding capacity) |
Huang et al. [39] |
|
Western Blotting |
Proteins separated by SDS-PAGE are transferred to membranes and detected with specific antibodies. |
Quantification of concentration and intensities of targeted proteins |
Zhang et al. [40] |
|
SWATH-MS (Sequential Window Acquisition of All Theoretical Mass Spectra) |
Sequential window acquisition fragments all precursor ions in small m/z windows, enabling reproducible DIA quantification. |
Helps establish protein libraries for meat quality control, safety, and traceability. |
López-Pedrouso et al. [41] |
Table 1: Current Proteomics Technologies Used in the Meat Industry.
Evolution of Proteomic Approaches
Early proteomic studies in meat science predominantly utilized two-Dimensional Gel Electrophoresis (2-DE) coupled with Mass Spectrometry (MS). This classic approach separates proteins by isoelectric point and molecular weight, followed by MS-based identification. While 2-DE remains valuable for resolving abundant and soluble proteins, it is limited in its ability to detect low-abundance, hydrophobic, or membrane-associated proteins [42]. The emergence of -omic technologies in poultry and other animal food research has further expanded the scope of proteomics, supporting the development of targeted therapeutic strategies and the detection of antibiotic resistance patterns [43].
To address the limitations of 2-DE, Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) has become the cornerstone of modern proteomics. LC-MS/MS offers superior sensitivity, dynamic range, and reproducibility, enabling the identification of thousands of proteins in a single analysis. It is compatible with both label-free and isobaric labeling strategies, such as isobaric Tags for Relative and Absolute Quantitation (iTRAQ) and Tandem Mass Tags (TMT), which facilitate multiplexed quantification and enhance throughput while minimizing variability [44,45].
Label-Free Quantification (LFQ) methods have also gained traction as cost-effective solutions for large-scale protein quantification, relying on spectral counting or peak intensity measurements. These approaches are particularly advantageous for studies requiring extensive sample comparisons without the constraints of chemical labeling. Recent innovations include Data-Independent Acquisition (DIA) techniques, such as SWATH-MS, which provide consistent and reproducible quantification across large sample sets and improve proteome coverage, making them highly suitable for biomarker discovery in meat science [20].
Collectively, these technological advancements have greatly expanded the depth and accuracy of protein analysis. They enable the detection of differential protein expressions linked to meat quality traits and facilitate the construction of protein interaction networks and pathway enrichment analyses. As a result, proteomic technologies are now indispensable for uncovering the molecular mechanisms underlying meat tenderness, color stability, lipid oxidation, and other quality-related attributes.
2-DE-Based Comparative Proteomics in Meat Science
Two-Dimensional Electrophoresis (2-DE) remains a foundational technique in comparative proteomics, widely applied in meat science for the separation and identification of proteins based on isoelectric point and molecular weight [46]. This method allows for the visualization of complex protein profiles and the comparison of differences between samples subjected to various conditions, such as species, breed, muscle type, storage, aging, and processing [47].
In the first dimension, proteins are separated by Isoelectric Focusing (IEF), and in the second dimension, by molecular weight using SDS-PAGE [48]. The resulting gel displays a pattern of protein spots, each corresponding to a unique protein or isoform. These spots can be excised, digested (commonly with trypsin), and identified by MS. 2-DE-based comparative proteomics is particularly effective for detecting Post-Translational Modifications (PTMs) and analyzing differential protein expression [49]. In meat science, this enables the study of biochemical changes related to tenderness, color stability, water-holding capacity, and oxidative stress [50]. For example, the abundance of enzymes such as calpains and cathepsins, which are associated with postmortem tenderization, can be monitored using 2-DE [51].
Additionally, 2-DE facilitates the discovery of biomarkers indicative of spoilage or quality deterioration, supporting improved meat safety and shelf-life prediction. When combined with image analysis software and statistical tools like Principal Component Analysis (PCA), 2-DE provides a robust framework for interpreting protein expression patterns across experimental groups [52]. Despite its strengths, 2-DE is less sensitive for hydrophobic and low-abundance proteins and struggles to resolve very large or small proteins [53,54]. Nevertheless, it remains a valuable method, especially when integrated with complementary approaches such as LC-MS/MS and bioinformatics analysis and continues to advance our understanding of meat quality at the molecular level.
DNA Barcoding and PCR for Species Authentication
Incidents of meat fraud, such as the 2013 horse meat scandal in Europe, have highlighted the critical role of DNA-based techniques in species authentication [55]. DNA barcoding and Polymerase Chain Reaction (PCR) have emerged as primary methods due to their high specificity, sensitivity, and robustness [56].
DNA barcoding involves amplifying and sequencing a standardized region of mitochondrial DNA-typically the cytochrome c oxidase I gene-which is conserved within species but variable between them [57]. The resulting sequence is compared against reference databases to determine species origin. PCR, especially real-time PCR (qPCR), enables rapid detection of species-specific DNA fragments using targeted primers and fluorescent probes [58]. These methods are highly effective even in processed or cooked meat products, where proteins may be degraded but DNA remains relatively intact [59]. DNA-based methods can detect trace levels of undeclared species, ensuring regulatory compliance and consumer trust. While proteomic techniques can also contribute to species identification through peptide profiling and MS [60], DNA-based approaches are considered the gold standard in forensic food analysis due to their accuracy, scalability, and legal admissibility [60].
Workflow of LC-MS/MS-Based Proteomic Analysis in Meat Samples
A typical workflow of figure 1, for protein analysis using LC-MS/MS in meat science begins with sample preparation, where meat tissue is homogenized under controlled conditions. Proteins are then extracted using appropriate lysis buffers to separate them from other cellular components. Following extraction, protein quantification and quality control are performed, often using colorimetric assays to determine concentration.
 Figure 1: The workflow of protein analysis.
Figure 1: The workflow of protein analysis.
Next, enzymatic digestion, commonly with trypsin, breaks down proteins into peptides. These peptides are separated by liquid chromatography and analyzed by tandem mass spectrometry, which identifies them based on mass-to-charge (m/z) ratios and fragmentation patterns. The resulting spectra are processed to determine peptide abundance, enabling the identification and quantification of proteins associated with key meat quality attributes. This workflow underpins the high-throughput, sensitive, and reproducible analysis that has become central to modern meat proteomics.
Application Of Proteomics In Quality Assessments
Proteomics has emerged as a powerful and versatile tool for assessing meat quality by providing detailed insights into the structural and functional changes of proteins during post-mortem aging, storage, and processing [61]. Protein degradation and modification are central to critical meat quality attributes such as tenderness, juiciness, water-holding capacity, and flavor development [62]. Proteomic approaches enable the identification of protein biomarkers associated with meat freshness, spoilage, oxidative stress, and technological treatments [63].
Post-mortem proteolysis, primarily mediated by calpains and cathepsins, plays a pivotal role in the development of meat tenderness [64]. Proteomic profiling has facilitated the monitoring of structural proteins such as troponin-T, titin, and desmin, which serve as indicators of meat aging and quality improvement over time [65]. For example, the degradation of troponin-T correlates with enhanced tenderness in aged beef [66]. The presence or absence of specific myofibrillar fragments can also indicate the aging status of meat, providing valuable information regarding consumer acceptability.
Spoilage induces significant proteomic alterations. Proteins susceptible to oxidation and microbial enzymatic activity can be detected using two-Dimensional Electrophoresis (2-DE) and mass spectrometry-based techniques [67]. For instance, increased protein carbonylation and depletion of sarcoplasmic enzymes such as enolase and creatine kinase have been linked to oxidative degradation during storage [68]. Studies in pork, chicken, and fish consistently demonstrate that protein oxidation-manifested by carbonyl formation or loss of sulfhydryl groups-correlates with reduced shelf life and diminished sensory quality [69]. Microbial spoilage further alters the muscle proteome through proteolytic degradation of actin, myosin, and metabolic enzymes, leading to off-flavors and texture deterioration [70]. Monitoring these protein changes enables the assessment of microbial contamination levels and differentiation between fresh and spoiled samples [71]. The accumulation of stress-related proteins, such as heat shock proteins, in spoiled meat further supports the use of proteomic markers for predicting quality [72].
Although proteomics provides valuable biochemical insights, no universal threshold for individual protein markers definitively indicates spoilage. Instead, the presence or depletion of specific protein fragments, combined with sensory evaluation and physicochemical parameters like pH and shear force, informs quality grading. Integrating proteomic data with sensory panels allows for the correlation of protein degradation products with consumer perceptions of freshness and palatability. This integrative approach holds promise for developing protein-based quality markers and more precise meat grading systems.
Species Authentication of Meat and Meat-Based Products via Proteomics
Species authentication is critical for ensuring the integrity, safety, and traceability of meat products [73]. While techniques such as Near-Infrared (NIR) spectroscopy are used, proteomics offers a chemically precise tool for species identification based on the unique structure and composition of proteins. Proteins differ between species in their amino acid sequences, post-translational modifications, and three-dimensional conformations, reflecting underlying genetic variations [74,75]. These differences manifest in species-specific peptides generated after enzymatic digestion (e.g., trypsin), forming unique proteomic signatures detectable by advanced analytical techniques [76].
The proteomics workflow for species authentication begins with protein extraction using buffers such as SDS or urea to solubilize proteins from meat samples. Enzymatic digestion produces smaller peptides that are separated by Liquid Chromatography (LC) and analyzed by Mass Spectrometry (MS), which measures their mass-to-charge ratios. Tandem MS (LC-MS/MS) further sequences peptides, enabling the identification of species-specific marker peptides. This combination of chemical separation and molecular identification underpins the accuracy and reliability of proteomics in species authentication. Duft et al. [77], illustrate this process in figure 2, demonstrating species identification in meat authentication.
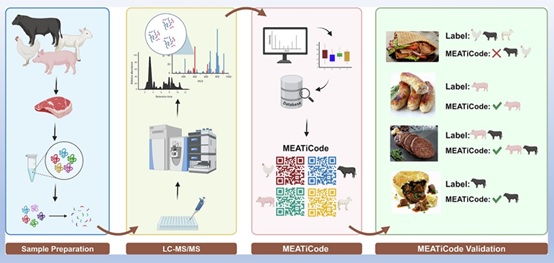 Figure 2: Meat species authentication techniques from processed food.
Figure 2: Meat species authentication techniques from processed food.
Species-specific peptides serve as chemical biomarkers due to their unique amino acid sequences, allowing differentiation even among closely related species such as sheep and goat or beef and buffalo [78,79]. Variations in peptide bond chemistry, amino acid side chains, and post-translational modifications like phosphorylation and oxidation contribute to these distinctive proteomic patterns [80].
Proteomics-based authentication offers high specificity and robustness, maintaining effectiveness in processed or cooked meats where DNA may be degraded. It enables both relative and absolute quantification of species content in complex mixtures, ensuring accurate labeling, consumer confidence, and regulatory compliance.
Proteomics in Meat Adulteration Detection
Proteomics has become a scientifically rigorous and analytically precise approach for detecting meat adulteration, particularly in processed and thermally treated products [81]. By comprehensively identifying and quantifying proteins, proteomics detects species-specific peptide markers that serve as molecular fingerprints of meat origin [16]. This is crucial when morphological features are lost, and DNA is fragmented due to processing [82].
Techniques such as LC-MS/MS and MALDI-TOF MS provide high-resolution peptide separation and characterization, ensuring sensitivity and specificity. Commonly targeted proteins include myosin, actin, and tropomyosin, whose peptide sequences differ between species and remain heat-stable [83]. Targeted proteomics strategies like Multiple Reaction Monitoring (MRM) enable quantification of trace amounts of undeclared meat with high confidence [84]. For example, detection of porcine-specific peptides in beef-labeled sausages unequivocally indicates adulteration, critical for consumer trust and compliance with religious and regulatory standards.
Proteomic data, matched with curated protein databases and validated by bioinformatics tools, allow accurate species identification in complex mixtures [24]. Although challenges such as high instrument costs and complex data interpretation persist, proteomics’ precision and versatility make it indispensable in modern meat authentication and food fraud prevention.
Challenges And Limitations Of Proteomics In Meat Research
Despite its transformative potential, the application of proteomics in meat research is accompanied by several significant challenges and limitations that can hinder its routine use in food authentication and quality control (Figure 3).
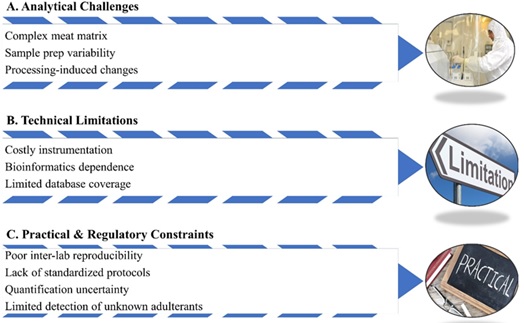 Figure 3: Challenges and the limitations of the proteomics study.
Figure 3: Challenges and the limitations of the proteomics study.
Complexity of the Meat Matrix
Meat is a highly complex biological matrix, comprising a diverse array of proteins, lipids, carbohydrates, and other biomolecules. This complexity can interfere with efficient protein extraction and identification, often necessitating extensive sample preparation steps such as solubilization, purification, and enzymatic digestion. These procedures are not only time-consuming but also susceptible to variability, which can impact reproducibility and data quality [85].
Effects of Processing and Storage
Thermal processing, curing, fermentation, and other technological treatments can induce protein denaturation, degradation, and chemical modifications. Such changes complicate the detection of intact species-specific peptides and may reduce the analytical sensitivity of proteomic methods. The altered protein landscape in processed meat poses additional challenges for accurate identification and quantification.
Instrumentation and Technical Expertise
Proteomics relies heavily on advanced, high-cost instrumentation such as high-resolution mass spectrometers (e.g., LC-MS/MS, MALDI-TOF). These instruments require substantial financial investment, regular maintenance, and highly trained personnel for operation and troubleshooting. The need for specialized infrastructure can limit accessibility, particularly in resource-constrained settings.
Bioinformatics and Database Limitations
The interpretation of proteomic data demands sophisticated bioinformatics tools and access to comprehensive, well-curated protein databases. However, existing databases may not cover all animal species or regional meat varieties, leading to potential false identifications or missed adulterants [86,87]. Incomplete or outdated databases can compromise the reliability of species authentication and adulteration detection.
Reproducibility and Standardization
Inter-laboratory reproducibility remains a persistent concern, as proteomic workflows are highly sensitive to variations in sample handling, instrument calibration, and data processing parameters. The lack of standardized protocols and validated methods for regulatory use further complicates the integration of proteomics into routine meat quality assurance [13].
Quantification and Detection of Adulterants
Quantifying adulterants using proteomics is challenging due to inherent differences in protein abundance across tissues and species. This variability can result in the underestimation or overestimation of adulteration levels, particularly in complex or highly processed products. Additionally, while proteomics is highly effective for detecting known species with established biomarkers, its capacity to identify unknown or unexpected adulterants is limited without prior reference data.
Future Directions
These challenges underscore the need for ongoing technological innovation, expansion and curation of protein databases, and the harmonization of analytical protocols. Addressing these limitations will be essential for the full integration of proteomics into mainstream meat authentication, quality control, and regulatory frameworks.
Conclusion
Proteomics has become a groundbreaking tool in meat science, providing precise and high-throughput analyses that exceed the limitations of traditional quality assessment methods. This technology allows for the identification and quantification of proteins, post-translational modifications, and degradation products, offering essential insights into meat quality traits, species authentication, and the detection of adulteration. Techniques such as LC-MS/MS, 4D label-free analysis, SWATH-MS, and TMT-based labeling (Table 1) have advanced the discovery of biomarkers related to tenderness, flavor, and oxidative stability while enhancing the reproducibility and robustness of quality evaluations. These methods are particularly valuable in addressing industry challenges, such as consumer trust, food fraud prevention, and regulatory compliance.
Despite these advancements, challenges persist, including the complexity of meat matrices, variability in processing-induced modifications, limitations in existing protein databases, and the need for standardized workflows across laboratories. Overcoming these challenges will necessitate continued innovation in mass spectrometry instrumentation, expansion of protein databases, and the development of harmonized bioinformatics platforms. Furthermore, combining proteomics with other omics technologies, such as metabolomics and volatilomics, shows promise for creating comprehensive predictive models of meat quality and safety.
In conclusion, the strategic application of advanced proteomic techniques in meat research will accelerate the development of reliable biomarkers, strengthen traceability systems, and improve routine quality assurance. By connecting molecular-level insights with industrial practices, proteomics will play a crucial role in ensuring meat authenticity, enhancing consumer confidence, and contributing to the overall sustainability of the global meat sector.
Highlights
- Proteomics ensures meat quality, authenticity, and adulteration detection.
- LC-MS/MS and SWATH-MS enable precise protein biomarker identification.
- Challenges include complex samples, protein loss, and costly instrumentation.
- Future focus: better databases, standardization, and omics integration.
Acknowledgement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ahamed Z, Saifullah M, Shellie R, Stanley RA (2025) Volatilomics in meat research: Species authentication, quality assessment & adulteration detection. Food Research International 214:
- Mia N, Alam A, Rahman MM, Ali MS, Hashem MA (2024) Probiotics to enhance animal production performance and meat quality: A review. Meat Research
- Sharker BC, Mia N, Ali MH, Hashem MA, Rahman MM, et al. (2024) Effect of freezing period on the quality of doe liver. Meat Research
- Hashem MA, Ambia J, Mia N, Ali MH, Rahman MM, et al. (2024) Detection of adulteration of goat and sheep meat through NIRS and chemometric analysis. Meat Research 4.
- Mia N, Sarker T, Halim MA, Alam A, Ali MS, et al. (2025) Machine learning overview and its application in the livestock industry. Meat Research
- Sarker T, Deen RA, Ghosh D, Mia N, Rahman MM, et al. (2024) AI driven approach and NIRS: A review on meat quality and safety. Meat Research
- Mia N, Rahman MM, Hashem MA (2023) Effect of heat stress on meat quality: A review. Meat Research
- Hashem MA, Sarker MSK, Alam AN, Mia N, Rahman MM (2024) Poultry processing and value addition in Bangladesh- A review. Meat Research
- Alam A, Mia N, Monti JA, Hashem MA, Ali MS (2024) Enhancing the qualitative attributes of meat through processing and preservation techniques- A review. Meat Research 4.
- Alam AN, Hashem M, Matar A, Ali M, Monti J, et al. (2024) Cutting edge technologies for the evaluation of plant-based food and meat quality: A comprehensive review. Meat Research
- Joo ST, Kim GD, Hwang YH, Ryu YC (2013) Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Science 95: 828-836.
- Vial C, Lamy A, Sebbane M (2025) Chefs saddle up-perceptions of horse meat as a sustainable gastronomic alternative in France. Foods 14:
- Martínez A, Abanto M, Días NB, Olate P, Nuñez IP, et al. (2025) Recent trends in food quality and authentication: The role of omics technologies in dairy and meat production. International Journal of Molecular Sciences 26:
- Banerjee R, Maheswarappa NB, Mohan K, Biswas S (2022) Proteomic approaches for the authentication of foods of animal origin. In Food Proteomics. Technological Advances, Current Applications and Future Perspectives 2022: 301-336.
- Sidira M, Agriopoulou S, Smaoui S, Varzakas T (2024) Omics-integrated approach (Metabolomics, Proteomics and Lipidomics) to assess the quality control of aquatic and seafood products. Applied Sciences 14:
- Banerjee R, Maheswarappa NB, Mohan K, Biswas S, Batabyal S (2022) Proteomic technologies and their application for ensuring meat quality, safety and Authenticity. Current Proteomics 19: 128-141.
- Jia W, Wu X (2023) Potential biomarkers analysis and protein internal mechanisms by cold plasma treatment: Is proteomics effective to elucidate protein-protein interaction network and biochemical pathway? Food Chemistry 426:
- Sukaew T (2024) The current and emerging research related aroma and flavor. In: Aroma and flavor in product development: Characterization, perception, and application. Springer Nature, Switzerland.
- Shakoor A, Zhang C, Xie J, Yang X (2022) Maillard reaction chemistry in formation of critical intermediates and flavour compounds and their antioxidant properties. Food Chemistry 393:
- Cao C, Xiao Z, Ge C, Wu Y (2022) Application and research progress of proteomics in chicken meat quality and identification: A review. Food Reviews International 38: 313-334.
- Kaur L, Hui SX, Morton JD, Kaur R, Chian FM, et al. (2021) Endogenous proteolytic systems and meat tenderness: Influence of post-mortem storage and processing. Food Science of Animal Resources 41:
- Cerqueira M, Magalhães CR, Schrama D, Farinha AP, Carrilho R, et al. (2024) Proteomics for comprehensive analysis of seafood products: Quality, safety, authentication, and allergen detection. In: Handbook of Seafood and Seafood Products Analysis. CRC Press, Florida, USA.
- Ewles M, Goodwin L (2011) Bioanalytical approaches to analyzing peptides and proteins by LC-MS/MS. Bioanalysis 3: 1379-1399.
- Chen C, Hou J, Tanner JJ, Cheng J (2020) Bioinformatics methods for mass spectrometry-based proteomics data analysis. International Journal of Molecular Sciences 21:
- Lytou AE, Panagou EZ, Nychas GJE (2019) Volatilomics for food quality and authentication. Current Opinion in Food Science 28: 88-95.
- Kuang L, He E, Zhou L, Lou A, Liu Y, et al. (2025) Dynamic changes in meat quality, volatile organic compounds, and microbial community of Xiangxi yellow cattle beef during chilled storage. Foods 14:
- Anwar AI, Ruslin M, Marlina E, Hasanuddin H (2025) Physicochemical analysis and application of sardinella fimbriata-derived hydroxyapatite in toothpaste formulations. BMC Oral Health 25:
- Yalew K, Pang X, Huang S, Zhang S, Yang X, et al. (2024) Recent development in detection and control of psychrotrophic bacteria in dairy production: Ensuring milk quality. Foods 13:
- Zhong R, Ji Z, Wang S, Chen H (2024) Bridging odorants and olfactory perception through machine learning: A review. Trends in Food Science & Technology 153:
- Adnane M, Almeida AM, Chapwanya A (2024) Unveiling the power of proteomics in advancing tropical animal health and production. Tropical Animal Health and Production 56:
- Seratlic S, Guha B, Moore S (2024) Advances in spectroscopic methods for predicting cheddar cheese maturity: A review of FT-IR, NIR, and NMR techniques. NDT 2: 392-416.
- Ayaz Q, Anjum N, Sidiq H, Naseem Z, Asad TS, et al. (2024) Structure, composition, nutritive value and biochemistry of post-mortem muscle. In: Hand Book of Processed Functional Meat Products. Springer Nature, Switzerland.
- Saini RV, Vaid P, Saini NK, Siwal SS, Gupta VK, et al. (2021) Recent advancements in the technologies detecting food spoiling agents. Journal of Functional Biomaterials 12:
- Xu J, Liu T, Luo R, Zheng C, Zhang Y, et al. (2025) Meat quality differences and protein molecular mechanisms affecting meat flavor in different breeds of Tibetan sheep analyzed by 4D label-free quantitative proteomics. Food Chemistry 480:
- Lin S, Sun Z, Li C, Kong B, Cao C, et al. (2025) Quantitative proteomics provides new insights into the mechanism underlying textural property improvement in frankfurters by ultrasound treatment combined with κ-ca Food Hydrocolloids 159: 110633.
- Zhang R, Xu M, Xu R, Bai T, Liu D, et al. (2025) Identification of Biomarkers for Meat Quality in Sichuan Goats Through 4D Label-Free Quantitative Proteomics. Animals 15:
- Sun X, He Z, Yang L, Wu H, Li H (2024) Quantitative proteomic analysis to identify potential biomarkers linked to quality traits of beef tripe from different sources. Food Chemistry 449:
- Govindaiah PM, Maheswarappa NB, Banerjee R, Mishra BP, Manohar BB, et al. (2023) Traditional halal meat production without stunning versus commercial slaughter with electrical stunning of slow-growing broiler chicken: Impact on meat quality and proteome changes. Poultry Science 102:
- Huang C, Blecker C, Chen L, Xiang C, Zheng X, et al. (2023) Integrating identification and targeted proteomics to discover the potential indicators of postmortem lamb meat quality. Meat Science 199:
- Zhang J, Toldrá F, Zhang W (2022) Insight into ultrasound-induced modifications of the proteome and flavor-related proteins of unsmoked bacon by applying label-free quantitation technology. Journal of Agricultural and Food Chemistry 70: 10259-10270.
- López-Pedrouso M, Lorenzo JM, Stasio LD, Brugiapaglia A, Franco D (2021) Quantitative proteomic analysis of beef tenderness of Piemontese young bulls by SWATH-MS. Food Chemistry 356:
- Zhang Z, Wu S, Stenoien DL, Paša-Tolic L (2014) High-throughput proteomics. Annual Review of Analytical Chemistry 7: 427-454.
- Tariq S, Samad A, Hamza M, Ahmer A, Muazzam A, et al. (2022). Salmonella in poultry; an overview. International Journal of Multidisciplinary Sciences and Arts 1: 80-84.
- Dias MH, Kitano ES, Zelanis A, Iwai LK (2016) Proteomics and drug discovery in cancer. Drug Discovery Today 21: 264-277.
- Chen L, Su W, Chen H, Chen DQ, Wang M, et al. (2018) Proteomics for biomarker identification and clinical application in kidney disease. Advances in Clinical Chemistry 85: 91-113.
- Murphy S, Dowling P, Ohlendieck K (2016) Comparative skeletal muscle proteomics using two-dimensional gel electrophoresis. Proteomes 4:
- Bouley J, Chambon, C, Picard B (2004) Mapping of bovine skeletal muscle proteins using two-dimensional gel electrophoresis and mass spectrometry. Proteomics 4: 1811-1824.
- Braun RJ, Kinkl N, Beer M, Ueffing M (2007) Two-dimensional electrophoresis of membrane proteins. Analytical and Bioanalytical Chemistry 389: 1033-1045.
- Ercan H, Resch U, Hsu F, Mitulovic G, Bileck A, et al. (2023) A practical and analytical comparative study of gel-based top-down and gel-free bottom-up proteomics including unbiased proteoform detection. Cells 12:
- Hollung K, Veiseth-Kent E (2012) Meat Science and Proteomics. In: OMICs Technologies: Tools for Food Science. Taylor & Francis, USA.
- Bischof G, Witte F, Terjung N, Heinz V, Juadjur A, et al. (2024) Metabolic, proteomic and microbial changes postmortem and during beef aging. Critical Reviews in Food Science and Nutrition 64: 1076-1109.
- Chich JF, David O, Villers F, Schaeffer B, Lutomski D, et al. (2007) Statistics for proteomics: experimental design and 2-DE differential analysis. Journal of Chromatography B 849: 261-272.
- Görg A, Drews O, Lück C, Weiland F, Weiss W (2009) 2-DE with IPGs. Electrophoresis 30: 122-132.
- Oliveira BM, Coorssen JR, Martins-de-Souza D (2014) 2DE: The phoenix of proteomics. Journal of Proteomics 104: 140-150.
- Bai JA, Rai VR (2017) Food fraud: Detection, prevention, and regulations. In: Food Safety and Protection. CRC Press, Florida, USA.
- Rahman MT, Uddin MS, Sultana R, Moue A, Setu M (2013) Polymerase chain reaction (PCR): A short review. Anwer Khan Modern Medical College Journal 4: 30-36.
- Roe AD, Sperling FA (2007) Patterns of evolution of mitochondrial cytochrome c oxidase I and II DNA and implications for DNA barcoding. Molecular Phylogenetics and evolution 44: 325-345.
- Hulley EN, Tharmalingam S, Zarnke A, Boreham DR (2019) Development and validation of probe-based multiplex real-time PCR assays for the rapid and accurate detection of freshwater fish species. PLoS One 14:
- Meyer R, Candrian U (1996) PCR-based DNA analysis for the identification and characterization of food components. LWT-Food Science and Technology 29: 1-9.
- Kumar P, Rani A, Singh S, Kumar A (2022) Recent advances on DNA and omics-based technology in Food testing and authentication: A review. Journal of Food Safety 42:
- Gagaoua M, Picard B (2022) Proteomics to explain and predict meat quality. In New aspects of meat quality. Woodhead Publishing Series in Food Science, Technology and Nutrition.
- Zhang H, Tang D, Yang H, Liu X, Cheng J, et al. (2023) Effects of high hydrostatic pressure assisted enzymatic tenderization on goose meat texture and myofibril protein. LWT-Food Science and Technology 184:
- Bassey AP, Ye K, Li C, Zhou G (2021) Transcriptomic-proteomic integration: A powerful synergy to elucidate the mechanisms of meat spoilage in the cold chain. Trends in Food Science & Technology 113: 12-25.
- Saccà E (2016) Expression of genes involved in meat tenderization of bovine and caprine muscles.
- Paredi G, Raboni S, Bendixen E, Almeida AM, Mozzarelli A (2012) “Muscle to meat” molecular events and technological transformations: The proteomics insight. Journal of Proteomics 75: 4275-4289.
- Jeneske H (2023) Accelerated aging affects shelf-stability, yield, sensory characteristics, cathepsin activity, and structure of collagen and troponin-T in lower quality beef cuts.
- Hadi MH, Dakhel SM, Jawad MS, Kazem RM, Abbas AS (2024) Microbial proteomics: Microbial stress adaptation, food safety, safety aspects of Genetically Modified Organisms [GMOs] and identification of foodborne pathogens and toxins. Current Clinical and Medical Education 2: 78-90.
- Piñeiro C, Martinez I (2012) Evaluation of fish quality and safety by proteomics techniques. In: Proteomics in Foods: Principles and Applications. Springer, Boston, USA.
- Zhang L, Li Q, Bao Y, Tan Y, Lametsch R, et al. (2024) Recent advances on characterization of protein oxidation in aquatic products: A comprehensive review. Critical Reviews in Food Science and Nutrition 64: 1572-1591.
- Prates JA (2025) Impact of heat stress on carcass traits, meat quality, and nutritional value in monogastric animals: Underlying mechanisms and nutritional mitigation strategies. Foods 14:
- Karanth S, Feng S, Patra D, Pradhan AK (2023) Linking microbial contamination to food spoilage and food waste: The role of smart packaging, spoilage risk assessments, and date labeling. Frontiers in Microbiology 14:
- Gagaoua M, Duffy G, Alvarez C, Burgess CM, Hamill R, et al. (2022) Current research and emerging tools to improve fresh red meat quality. Irish Journal of Agricultural and Food Research 61: 145-167.
- Vishnuraj MR, Kumar NA, Vaithiyanathan S, Barbuddhe SB (2023) Authentication issues in foods of animal origin and advanced molecular techniques for identification and vulnerability assessment. Trends in Food Science & Technology 138: 164-177.
- Hudson LD, Nadon NL (2023) Amino acid substitutions in proteolipid protein that cause dysmyelination. In: Myelin. Routledge, UK.
- Bradley D (2022) The evolution of post-translational modifications. Current Opinion in Genetics & Development 76:
- Stachniuk A, Sumara A, Montowska M, Fornal E (2021) Liquid chromatography-mass spectrometry bottom-up proteomic methods in animal species analysis of processed meat for food authentication and the detection of adulterations. Mass Spectrometry Reviews 40: 3-30.
- Duft RG, Griffin JL, Stead DA (2025) MEATiCode: A comprehensive proteomic LC-MS/MS method for simultaneous species identification in meat authentication. Food Chemistry 483:
- Gu Y, Zhang J, Sun J, Yu H, Feng R, et al. (2021) Marker peptide screening and species-specific authentication of Pheretima using proteomics. Analytical and Bioanalytical Chemistry 413: 3167-3176.
- Chaudhary P, Kumar Y (2022) Recent advances in multiplex molecular techniques for meat species identification. Journal of Food Composition and Analysis 110:
- Kumar A, Narayanan V, Sekhar A (2019) Characterizing post-translational modifications and their effects on protein conformation using NMR spectroscopy. Biochemistry 59: 57-73.
- Li YC, Liu SY, Meng FB, Liu DY, Zhang Y, et al. (2020) Comparative review and the recent progress in detection technologies of meat product adulteration. Comprehensive Reviews in Food Science and Food Safety 19: 2256-2296.
- Nehal N, Choudhary B, Nagpure A, Gupta RK (2021) DNA barcoding: A modern age tool for detection of adulteration in food. Critical Reviews in Biotechnology 41: 767-791.
- Chhikara A, Kumari P, Dalal J, Kumari K (2024) Protein degradation patterns as biomarkers for post-mortem interval estimation: A comprehensive review of proteomic approaches in forensic science. Journal of Proteomics 310:
- Agregán R, Pateiro M, Kumar M, Franco D, Capanoglu E, et al. (2023) The potential of proteomics in the study of processed meat products. Journal of Proteomics 270:
- Nawaz A, Irshad S, Khan IA, Khalifa I, Walayat N, et al. (2022) Protein oxidation in muscle-based products: Effects on physicochemical properties, quality concerns, and challenges to food industry. Food Research International 157:
- Ortea I, O’Connor G, Maquet A (2020) Review on proteomics for food authentication. In: Proteomics for Food Authentication. CRC Press, Florida, USA.
- Ramanathan R, Kiyimba F, Suman SP, Mafi GG (2023) The potential of metabolomics in meat science: Current applications, trends, and challenges. Journal of Proteomics 283: 104926.
Citation: Tariq S, Mia N, Ahamed Z (2025) Proteomics Methods in Meat Research: A Review of Applications for Species Authentication, Quality Evaluation, and Adulteration Detection. HSOA J Food Sci Nutr 11: 229.
Copyright: © 2025 Sania Tariq, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

