
The Exploration of Age Related Mirna in Saliva Based On Massively Parellel Sequencing: A Pilot Study
*Corresponding Author(s):
Chen FangSchool Of Basic And Clinical Medicine, Shandong First Medical University& Shandong Academy Of Medical Sciences, Jinan 250117, China
Email:chenfangyulix@126.com
Song Zhou
Jinan Central Hospital, Shandong First Medical University & Shandong Academy Of Medical Sciences, Jinan, Shandong, 250013, China
Email:woshizhousong@126.com
Abstract
Age determination is one of the key components of forensic science research. Accurate age judgment can provide crucial leads for case-solving and holds significant importance. The methods used for age determination in forensic cases, such as the bone age and dental age estimation mainly based on morphology, the methods for age determination from bodily fluids such as saliva and blood mainly based on DNA methylation, however, the process is complex and the accuracy is insufficient. In this study, saliva miRNA expression profiles of male population were analyzed based on massively parallel sequencing, age-related miRNAs were screened based on massively parellel sequencing. According to our results, three miRNAs miR-128-3p, miR-125a-5p and miR-125b-5p were down-regulated with aging and two miRNAs miR-142-3p, miR-486-3p were up-regulated with aging. Multiple machine learning methods were applied in the age classification model.
AdaBoost algorithm resulted in the highest accuracy (Precision) of the age inference model, 66.7% samples were accurately classified. This pilot study indicated that miRNAs may provide novel biomarkers to address the forensic challenge of saliva-based age determination.
Keywords
Age estimation; microRNA; Saliva
Introduction
Age inference holds significant importance in forensic identification. Commonly applied methods for age inference based on the valuating the maturity of the skeleton, dental development. Recently, the age estimation of trace samples, such as blood and saliva are widely focused upon by people. Saliva is a special bodily fluid secreted by various salivary glands,as a common type of biological sample, the collection of saliva has the advantages of being fast, simple, and non-invasive [1], and with the rapid development of molecular biology, various biomarkers such as RNA, DNA, and proteins in saliva have also been utilized. Therefore, extracting extensive information from saliva stains has the potential to significantly advance the resolution of forensic investigations. Saliva residue, often recoverable from bite marks on a victim's skin or traces found in more intimate areas, can be instrumental in reconstructing the sequence of events, potentially indicating the nature of a crime, such as a sexual assault [2]. Recent efforts to estimate the age of saliva stains have largely centered on DNA methylation [3,4]. Nonetheless, current DNA methylation techniques have notable constraints, including procedural complexity and limited accuracy, pointing to a need for the discovery of additional biomarkers to enhance the age estimation of saliva in forensic science.
MicroRNAs (miRNAs) are endogenous, single-stranded non-coding RNAs, typically comprising 18 to 25 nucleotides, that play diverse roles in biological function. These molecules function analogously to transcription factors, offering benefits such as compact molecular structure, considerable stability, and tissue-specific expression profiles. MiRNAs are capable of modulating an array of biological mechanisms, including cell proliferation, tumorigenesis, and cellular differentiation. This regulatory capacity implies that miRNAs may possess inherent specificity for discerning unique molecular signatures [5,6]. The previous studies showed that miRNAs were widely expressed in saliva and may function as a promising tool for future biomarker studies [7].
Due to the temporal nature, tissue specific expression, and stability of miRNA expression, there is a vast potential for exploration in the field of age determination for forensic evidence [8]. Studies by Nicole and others have shown that through fluorescent quantitative PCR methods, the miRNA expression levels in peripheral blood mononuclear cells of people of different ages were analyzed, finding that with increasing age, miRNAs such as miR-103 exhibited a noticeable downward trend [9]. Our group detected that both piRNAs and miRNAs have potential in the age estimation. Thus, differential expression of miRNAs can be utilized in research for age determination methods. In this study, the age related miRNAs in healthy male saliva were detected based on massively parellel sequencing and the age classification model was established using machine learning method. Our work will provide new direction of age estimation for body fluid.
Materials and Methods
- Collection of saliva samples
Eighteen saliva samples were collected from healthy male volunteers in Shandong province and written informed consent was obtained. The Body Mass Index of the volunteers ranged from 23 to 27. There are three ages groups: 20, 35, 50. Each age group has six samples. All experiments were carried out in accordance with the guidelines and regulations of the Ethical Committee of Shandong First Medical University. The committee assessed and approved the study protocol.
- MiRNA isolation and quantification
MiRNA was extracted from saliva samples using the mirVana™ PARIS™ RNA and Native Protein Purification Kit (Ambion, USA). For saliva samples, 600 μL saliva was applied in the experiment. The saliva was mixed with ice-cold 600 μL disruption buffer for 5 min. Then the supernatants were transferred to a new tube and mixed with 400 μL denaturing solution at room temperature (22-24°C). RNA was dissolved in 20μL RNase free water and the purity and quantity of RNA were assessed using NanoDrop ND-1000 spectrophotometer (Thermo Scientific, USA). RNA integrity was assessed using the RNA Nano 6000 assay kit (Agilent, USA) of the Agilent Bioanalyzer 2100 system (Agilent Technologies, USA). The detail information of the integrity and concentration of RNA was provided in the supplementary table S1. The obtained RNA was stored at 80°C until use.
- MiRNA sequencing
Total RNA (100 ng) per sample was used as the input material generating the small RNA library. Sequencing were performed in two flow cells, separately. Sequencing libraries were generated using NEBNext® multiplex small RNA library prep set for Illumina® (7300L, New England Biolabs, USA) according to the manufacturer’s recommendations. After the 3´SR and 5´ SR adaptors were ligated, reverse transcription was performed and the index codes were added to attribute unique barcodes to each sample. Next we purified the PCR amplifed cDNA construct using a QIAQuick PCR purifcation kit (QIAGEN, German). Samples were loaded on the 6% poly acrylamide gel (Thermo Fisher, USA) and electrophoresed for 1h at 120V or until the blue dye reached the bottom of the gel. The 140 and 150 nucleotide bands corresponded to adapter-ligated constructs derived from the 21 and 30 nucleotide RNA fragments, respectively. Bands corresponding to 148 bp were isolated. Finally, libraries were dissolved in 12 µL Tris-EDTA buffer. Library quality was assessed on the Agilent Bioanalyzer 2100 system using DNA high sensitivity chips (Agilent, USA). The concentration was detected using the KAPA library quantification kit (KAPA, USA). Library preparations were sequenced on an Illumina Hiseq 2500 platform (Illumina, USA) and 50-bp single-end reads were generated. From the remaining sequences, only those that were 15–27 bases long were retained for further analysis because they most likely contained mature miRNA sequences. The miRDeep2 software (MDC, Germany) was applied to identify miRNAs. MiRDeep2 was run on the data with default options. Known miRNA inputs were from the miRBase version 21. The miRDeep2 program was used to identify novel miRNAs in a sequencing dataset. All mapped reads were normalized to reads per million.
- Data analysis
All data are presented as mean ± standard error of mean. The Kolmogorov-Smirnov test was performed to determine whether the data were normally distributed. Two-tailed, independent sample t-tests or variance (ANOVA) with LSD post hoc tests were used and p < 0.05 was considered as statistically significant. Statistical analysis was performed using the statistical software program package Prism 5 (GraphPad, USA). The R packages edge R and limma were implemented to analyze the differential expression of miRNA expression levels determined from sequencing. Heatmap analysis was conducted using HemI (Cuckoo group, China, URL: www.hemi.biocuckoo.org).
- Establishment of machine learning models
For model fitting, the whole RNA dataset from the saliva samples was applied in the establish the age classification model. Using the dataset, Orange software (Orange, Slovenia) was applied to assess the different models.
Results
The expression profile of miRNAs in saliva
A total of 141 miRNAs were detected based on massively parellel sequencing. Compared with the 20-year-old group, 58 miRNAs increased and 68 miRNAs decreased in the 35-year-old group. Compared with the 50-year-old group, 55 miRNAs increased and 50 miRNAs decreased in the 35-year-old group (Figure 1).
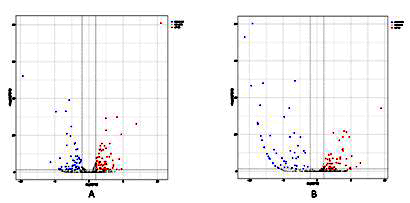 Figure 1: The miRNAs with differences with aging.A. The volcano plot of differentially expressed miRNAs between 20-year-old age group and 35-year-old age group. B. The volcano plot of differentially expressed miRNAs between 35-year-old age group and 20-year-old age group. The blue dots indicates the miRNAs is reduced and the red dots indicates the miRNAs is increased.
Figure 1: The miRNAs with differences with aging.A. The volcano plot of differentially expressed miRNAs between 20-year-old age group and 35-year-old age group. B. The volcano plot of differentially expressed miRNAs between 35-year-old age group and 20-year-old age group. The blue dots indicates the miRNAs is reduced and the red dots indicates the miRNAs is increased.
The age related miRNAs in saliva
From the massive parellel sequencing results, we found that the expression of hsa-miR-128-3p, hsa-miR-125a-5p, hsa-miR-125b-5p was down regulated with aging, compared with the 20-year-old age group, the expression level of each miRNA reduced 66.43%, 84.52% and 69.42% in 50-year-old age group, while the expression of hsa-miR-142-3p, hsa-miR-486-3p was up regulated with aging, compared with the 20-year-old age group, the expression level of each miRNA increased eight point three-fold and thirteen point five-fold in 50-year-old age group(Figure 2 and 3).
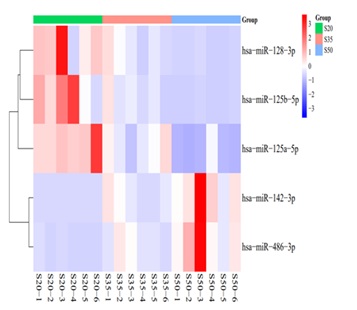 Figure 2: The heat map results showed that the expression of hsa-miR-128-3p, hsa-miR-125a-5p, hsa-miR-125b-5p was down regulated, while the expression of hsa-miR-142-3p, hsa-miR-486-3p was up regulated.(S20: the saliva from the 20-year-old age group, S35: the saliva from 35-year-old age group, S50 the saliva from the 50-year-old age group).
Figure 2: The heat map results showed that the expression of hsa-miR-128-3p, hsa-miR-125a-5p, hsa-miR-125b-5p was down regulated, while the expression of hsa-miR-142-3p, hsa-miR-486-3p was up regulated.(S20: the saliva from the 20-year-old age group, S35: the saliva from 35-year-old age group, S50 the saliva from the 50-year-old age group).
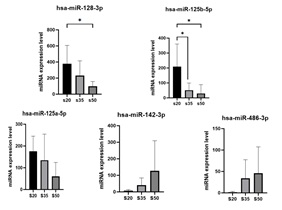 Figure 3: The expression of hsa-miR-128-3p, hsa-miR-125a-5p, hsa-miR-125b-5p hsa-miR-142-3p and hsa-miR-486-3p in different age group. (n=6, *p < 0.05, **p < 0.01)
Figure 3: The expression of hsa-miR-128-3p, hsa-miR-125a-5p, hsa-miR-125b-5p hsa-miR-142-3p and hsa-miR-486-3p in different age group. (n=6, *p < 0.05, **p < 0.01)
Age classification model based on machine learning
Using Orange software, age classification models were established using three miRNA, hsa-miR-128-3p, hsa-miR-125a-5p, hsa-miR-125b-5p which were negatively correlated with age (Table 1). SVM, kNN, AdaBoost, Naive Bayes, logistic regression, and random forest algorithms were used to evaluate the models. The results showed that the AdaBoost algorithm resulted in the highest accuracy (Precision) of the age inference model, 66.7% samples were accurately classified. Next are logistic regression and Naive Bayes, with accuracies of 0.621 and 0.660.
|
Model |
AUC |
CA |
F1 |
Precision |
Recall |
|
Naive Bayes |
0.745 |
0.611 |
0.613 |
0.621 |
0.611 |
|
Logistic Regression |
0.759 |
0.667 |
0.660 |
0.660 |
0.667 |
|
kNN |
0.576 |
0.444 |
0.459 |
0.483 |
0.444 |
|
AdaBoost |
0.667 |
0.556 |
0.571 |
0.667 |
0.556 |
|
SVM |
0.602 |
0.444 |
0.443 |
0.446 |
0.444 |
|
Random Forest |
0.704 |
0.444 |
0.446 |
0.451 |
0.444 |
Table 1: The age classification model based on machine learning method.
Discussion
Saliva stain have always been a significant presence in forensic research. They encompass various aspects. Research has shown that by detecting the ratio of residual urea concentration to amino acid concentration in saliva plaques, it has been found that it fluctuates regularly over time. It can be inferred whether it occurred before or after the case at a certain residual time, which also provides new clues for judging the case [10]. And there is often more than one type of plaque in crime scenes, and with the discovery of various biomarkers, establishing a complete age inference model based on different biomarkers has become a direction that can be continuously deepened. And today's DNA methylation is like this. From 1973, Vanyushin et al. found that the overall level of DNA methylation changes with age in rats through research [11]. To now, studies have shown that through DNA methylation, more accurate age inference models suitable for blood, blood stains, saliva stains, etc. can be accurately established, but it has a significant bias in predicting muscle tissue, even exceeding the age of 10 years [12]. At the same time, DNA methylation analysis has cumbersome steps and high requirements for sample quality, which urgently requires the establishment of new age inference methods. In our previous study, we have detected piRNAs and miRNAs have potential in age estimation in blood samples [13]. But the studies focused on saliva age estimation using miRNAs are still lacking.
In this experiment, high-throughput sequencing, we screened for age related differentially expressed miRNAs that can be used for saliva spot analysis in male samples in China. In our study, hsa-miR-128-3p, hsa-miR-125a-5p and hsa-miR-125b-5p, were found to be down regulated with age, two miRNAs, hsa-miR-142-3p and hsa-miR-486-3p were up regulated with age. The age related miRNAs found in this study also have great potential in clinical research. Studies have shown that hsa-miR-125a-5p can serve as a potential biomarker in the diagnosis of severe depression and bipolar disorder [14]. And hsa-miR-128-3p can be applied in early screening of high-risk breast benign tumors [15]. This pilot study first established the age classification model in saliva using miRNAs. The shortcoming of the research is the results has not been validated using forensic on-site samples such as saliva stains. Moreover, this study also suffers from an issue of insufficient sample size. In the next step of research, the sample size will be further increased, the age inference model will be optimized, and the established method will be applied in samples from forensic cases, such as saliva stain.
Conclusion
This pilot study employs high-throughput sequencing technology to examine the salivary miRNA expression profiles across various age groups within the Han Chinese population. By establishing an age estimation method predicated on salivary miRNA, it offers novel perspectives for tackling the complexities of age prediction in forensic biological stain analysis.
Declarations
- Ethics approval
The study was Ethics Committee of Shandong First Medical University (approval number 2022-729).
Conflict of Interest
The authors declare that they have no conflict of interest.
Acknowledgement
This work was supported by National Natural Science Foundation of China (No. 82002006), the Open project of Shanghai Key Laboratory of Forensic Medicine, Key lab of Forensic Science, Ministry of Justice, China (Academy of Forensic Science) (KF202206) and Qingchuang Talents Induction Program of Shandong Higher Education Institution (2022 Forensic medicine innovation team).
Reference
- Du X, Liang X, Zhou X (2023) Research progress on saliva biomarkers in the diagnosis of systemic diseases. Journal of Sichuan University (Medical Science 54: 33-38.
- Bond JW, Hammond C (2008) The value of DNA material recovered from crime scenes. Forensic Sci 53: 797-801.
- Fokias K, Dierckx L, Van de Voorde W (2023) Age determination through DNA methylation patterns in fingernails and toenails. Forensic Sci Int Genet 64: 102846.
- Jung SE, Lim SM, Hong SR, Lee EH, Lee HY, et al. (2019) DNA methylation of the ELOVL2, FHL2, KLF14, C1orf132/MIR29B2C, and TRIM59 genes for age prediction from blood, saliva, and buccal swab samples. Forensic Sci Int Genet 38: 1-8.
- Fang C, Zhao J, Li J, Qian J, Liu W, et al. (2019) Massively parallel sequencing of microRNA in bloodstains and evaluation of environmental inflfluences on miRNA candidates using realtime polymerase chain reaction. Forensic Sci Int Genet 38: 32-38.
- Silva SS, Lopes C, Teixeira AL (2015) Forensic miRNA: Potential biomarker for body flfluids? Forensic Sci Int Genet 14: 1-10.
- Hofmann L, Kors TA, Ezic J, Niesler B, Röth R, et al. (2022) Comparison of plasma- and saliva-derived exosomal miRNA profiles reveals diagnostic potential in head and neck cancer. Front Cell Dev Biol 10: 971596.
- Hu R, Fang C, Liu X (2016) Advances in miRNA analysis in forensics. Chinese Journal of Forensic Medicine 31: 456-462.
- Tian H, Lv M, Li Z, Peng D, Tan Y, et al. (2017) Semen-specific miRNAs: Suitable for the distinction of infertile semen in the body fluid identification. Forensic Sci Int Genet 33: 161-167.
- Quan B, Li S, Cheng Y (2019) An experimental study to extrapolate the time of saliva plaque retention . Chinese Journal of Forensic Medicine 34: 539-544.
- Vanyushin BF, nemirovsky LE, klimenko VV (1973) The 5-methylcytosine in DNA of rats.Tissue and age specificity and the changes induced by hydrocortisone and other agents. J Gerontologia 19: 138-152.
- Jia F, Sun X, Yu S, et al. (2021) Forensic individual age inference of DNA methylation levels. Chinese Journal of Forensic Medicine 36: 373-378.
- Fang C, Zhou P, Li R, Guo J, Qiu H, et al. (2023) Development of a novel forensic age estimation strategy for aged blood samples by combining piRNA and miRNA markers. Int J Legal Med 137: 1327-1335.
- Gecys D, Dambrauskiene K, Simonyte S, Patamsyte V, Vilkeviciute A, et al. (2022) Circulating hsa-let-7e-5p and hsa-miR-125a-5p as Possible Biomarkers in the Diagnosis of Major Depression and Bipolar Disorders. Dis Markers 2022: 3004338.
- Khadka VS, Nasu M, Deng Y (2023) Circulating microRNA Biomarker for Detecting Breast Cancer in High-Risk Benign Breast Tumors. Int J Mol Sci 24: 7553.
Citation: Guan Z, Zhao C, Wu J, Li M, Zhang Q, et al. (2024) The Exploration of Age Related Mirna in Saliva Based On Massively Parellel Sequencing: A Pilot Study. Forensic Leg Investig Sci 10: 091.
Copyright: © 2024 Zimeng Guan, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

