
The Potential of Distinct Species of Non-Human Model Organisms for Research of Neurodegenerative Diseases
*Corresponding Author(s):
Xiucheng Eric ZhangBlair Academy, 2 Park St, Blairstown, NJ 07825, United States
Email:zhangx@blair.edu
Abstract
Neurodegenerative diseases (NDDs) such as Alzheimer’s Disease and Parkinson’s Disease are becoming increasingly prevalent. There is an urgent need to develop new and more effective therapeutic strategies to combat these devastating illnesses. However, due to ethical concerns regarding experimentation on human beings to research NDDs, little progress has been made with human research subjects. As a result, model organisms have become our best approach for better understanding the underlying mechanisms behind NDDs. This research paper aims to explore the potential candidates for model organisms in order to determine what species would have the greatest potential to study human NDDs and potentially allow for more effective research for therapies in the future. In order to find the ideal model organism, we conducted a thorough analysis that compiled data from various sources. The method in which the best model organism was determined utilized genotypic and phenotypic comparisons between the model organisms, as well as genetic associations made between these model organisms and a set of characteristic NDDs. Moreover, we provide a list of the advantages and limitations of many commonly used model organisms. In conjunction with our qualitative comparisons of genetic association and phenotypic effects of the animal models, we provided quantitative comparisons that included several examples of life history data. These groups of data were then integrated and analyzed together, and our results suggest that a certain species of model organism is better suited for the study of NDDs than others. Based on the findings, it would be sensible to invest more effort into researching neurodegenerative diseases using this standard model organism to increase the efficacy of scientific modeling and research.
Keywords
Alzheimer’s disease; Parkinson’s disease; Neurodegenerative diseases
INTRODUCTION
Neurodegenerative disorders (NDDs) are characterized by their detrimental effects on neurons. Since these cells are the primary components of the nervous system and do not replicate by mitosis, these diseases cause a myriad of neurological and physical impairments [1]. Some of the prevalent NDDs include: Parkinson’s Disease (PD), Alzheimer's Disease (AD), and Frontotemporal Dementia (FTLD). The risk of developing these disorders and other similar diseases has a strong positive correlation with age. Research has shown the connection between genetic architecture and the development of these NDDs. For example, harboring the apolipoprotein E4 (APOE4) allele is a major risk factor in developing early onset AD, and is associated with the loss of neurological synapses and the buildup of beta-amyloid plaques between neurons [2]. This damage causes the symptoms of Alzheimer’s, which include memory loss, difficulty communicating, and cognitive impairment.
FTLD, also known as Pick’s Disease, is a form of dementia resulting from damage caused to the frontal and temporal lobes; it thus causes changes to a patient’s emotions and language skills. A gene that has been studied relating to this is the Tau or MAPT gene, which causes tau protein abnormalities. Additionally, candidate gene and Genome Wide Association Studies (GWAS) have unearthed several genes and mutations that are associated with monogenic PD, such as the PARK8 autosomal-dominant allele [3]. As a result, these alleles are strongly correlated with the effects of PD, which include cognitive impairment, dementia and tremors.
For most NDD cases, the genotype of the individual is a major factor in their development of symptoms. However, there are certain environmental influences that can catalyze the progression of disorders. The development of PD, for instance, may be impelled by chemical compounds such as MPTP, which destroy neurons that transmit dopamine, a neurotransmitter related to movement. Thus, the behavioral and physical effects of the disease are intensified. Industrial gases and certain pharmaceutical products have a part to play in the advancement of PD [4]. In addition to these, there are other factors that have been observed to be related to the cause of PD, such as ingesting pesticides, whether through food or water [5]. These NDDs have devastating impacts on the quality of life of sufferers. For example, as PD progresses with age, it causes muscle degeneration, loss of vocal abilities, and diminished bodily function control. The very nature of these diseases causes patients to not have an understanding of what is happening to them; though the changes to their body are physical, they cannot see and thus know what exactly is occurring inside their brains. Additionally, those who suffer from NDDs often require intensive care, which is quite time-consuming [6,7] and expensive if being conducted through an outside organization [8].
Experimentation on human beings is viewed as unethical, making it difficult to conduct research on NDDs that affect humans. However, model organisms present less ethical concerns, and have already changed the study of human biology and disease pathology in ways that were not predictable a decade ago; model organism studies could help circumvent the ethical concerns of experimentation on humans. With animal models of NDDs, knowledge and understanding of the molecular pathogenesis of neurodegenerative disorders such as Alzheimer’s Disease, Parkinson’s Disease, and Frontotemporal Dementia has been enhanced. Once a causal allele has been associated with a disease phenotype, scientists can create knockout (KO) or transgenic animals in order to better understand the mechanisms of these diseases. In addition to bypassing concerns on ethics, the use of model organisms to study human diseases serves as a convenient method to analyze many disorders like AD, FTLD, and PD. A myriad of organisms have been used to study human diseases, including mice, monkeys, fruit flies, zebrafish, and certain types of nematodes [9]. Model organisms are viable, ethical research subjects that can be utilized to study the underlying causes and effects of the many components behind these diseases. However, model organism studies of NDDs can still be improved. Currently, a variety of model organisms are used to study NDDs, and it is unclear if any are more suitable to model them. This research aims to determine the best model for improved understanding and treatment of these neurological diseases.
In order to determine which model organisms should be used for the study of NDDs, we compare them in respect to their genetic similarity to humans and the overarching characteristics that compose a model organism (reproduction rate, generation time, etc). We also review literature using model organisms to study NDDs, measuring the overall success of each model organism for a range of diseases in terms of number of genetic associations made as well as their phenotypic effects. Our ultimate goal is to dictate the best model for the study of NDDs to improve our ability to conduct more efficient research on these disorders.
We hypothesize that the rhesus monkey (macaca mulatta) is the best model organism for the study of neurodegenerative diseases because of its genetic similarity to humans. The rhesus monkey is classified as a mammal, which indicates close characteristics to humans. Furthermore, rhesus monkeys share a common ancestor with humans approximately 8 million years ago, which is relatively recent, in terms of evolution. Additionally, rhesus monkeys and other primates exhibit behavioral characteristics comparable to humans. Taking into consideration the fact that most neurodegenerative diseases have phenotypic effects on behavioral traits, the idea that similar behavior may increase the potential of the model organism is plausible.
METHODS
To conduct our research, we performed a meta-analysis that consolidated sets of data from a myriad of academic sources. This data was then statistically analyzed in order to determine whether our hypothesis was to be accepted or rejected. To compile our data, we utilized various literature reviews and articles available through the University of California, Santa Barbara Library’s research database. During the search, a certain criterion of search words were used to efficiently sort through the documents. Keywords included: Model Organism, Genome, Gene-Editing, Neurodegenerative Disease, Parkinson’s Disease, Non-human model organism, Schizophrenia, Statistical Analysis, Alzheimer’s, Transgenic systems, Frontotemporal Dementia, and APOE4. When collecting information from these articles and reviews, we categorized the data into two distinct groups: the general characteristics of a model organism, and specific observations made by scientists in previous studies and experiments. This particular organization of data reflects the two main spheres of influence we need to consider when identifying the ideal organism for modeling NDDs.
First, there are certain characteristics that an organism must have in order to make it ideal for utilization in a lab, regardless of the topic being researched. This category of information is presented as general characteristics of a model organism, and includes life history traits (for example, generation and maturation times) and certain neurological attributes, such as the number of neurons in the brain.
The second category of information is centered on the data collected by various scientists studying each model organism and their overall success (if any) with the organism in their research. For this group of data, we viewed the genetic associations of the NDD, and the phenotypes that the model organism displayed when the NDD was introduced to its nervous system. By genetic associations, we mean the number of genes affecting a particular disease. We also created a list of the benefits and limitations that each model organism presented with every disease introduced.
The NDDs listed in our study were chosen for distinct reasons. The three NDDs we picked share genetic association in the sense that the Tau protein is a cause for all three diseases. In addition, PD is one of the most common NDDs and the most physically degrading. AZ is very well-known in popular science and patients of this suffer from severe memory loss.10 Lastly, FTLD is a type of dementia that accounts for 20-50% of all dementia cases; it also defines a group of disorders with similar characteristics, and this breadth allows for our findings to have a greater significance and impact on the human population. Through our comparative analysis of these two data sets, we were able to design a method that determines the ideal organism for modeling NDDs.
RESULTS
Model organisms are generally selected for a rapid generation time and substantial number of offspring, among other things. All of the candidate species have differing benefits and limitations in reference to their life history traits. These qualities often determine whether an organism can even be considered for use. Furthermore, other qualifications vary, depending on what disease the target organism will study. The parameters of an ideal organism for modeling NDDs typically include the genetic resemblance to humans who suffer from the disease as well as phenotypic similarity of effects.
General characteristics
When looking at overall attributes of model organisms, mice appear to have moderate statistics in comparison to the other four species (Table 1). In all three categories, it possesses qualities that hover around the mean. The fruit fly has a relatively short generation time of around 12 days, and synthesizes the most offspring of the five species. Roundworms have a short generation time of four days and produce a large amount of offspring. Zebrafish are similar to mice in generation maturation time, but produce a substantially greater quantity of offspring. The rhesus monkey, however, appears to be an outlier in the group. It has the longest generation time, slowest reproductive maturity, and gives birth to the least amount of offspring. In a lab, obtaining F1 and F2 generations would be time-consuming when viewed with the other organisms. All these factors are taken into consideration when deciding which organism would act as an efficient model.
|
Species of Model Organism |
Generation Time (months) |
Amount of Offspring per Reproductive Event |
Time to Reach Maturity (months) |
References |
|
Mouse Mus musculus |
3 |
6-8 |
1.5-2 |
S. Graham, M. Nei, S. Herculano-Houzel, R. Lambert, L. Ballanger |
|
Fruit fly Drosophila melanogaster |
12/30 |
500 |
10/30 |
G. Rubin, J. Shih, H. Lagercrantz, P. Geiger, T. Wong |
|
Roundworm Caenorhabditis elegans |
4/30 |
300 |
3/30 |
S. Perry, O. Hobert, D. Muschiol, L. Herndon, J. Chasnov, R. Wilson |
|
Zebrafish Danio rerio |
3-4 |
100-200 |
3 |
K. Hinsch, J. Postlethwait, C. Lawrence, Sanger Institute, D. Ho, C. Singleman |
|
Rhesus monkey Macaca mulatta |
132 |
1 |
36-48 |
C. Xue, C. Choi, S. Herculano-Houzel, NIH, M. Kessler |
Table 1: The table refers to the different life history traits of each model organism. A quick generation time, high count of off spring, and short period until sexual maturity are all favored general characteristics of an ideal model organism.
In addition, we modeled our general understanding of each model organism in a bar graph (Figure 1). The percentage of the genome sequenced was used as a measurement of our current understanding of the organism. All five organisms have a majority of their genomes sequenced, but only the zebrafish and rhesus monkey have their genomes completely sequenced. Meanwhile, the other three organisms still have the bulk of their DNA sequenced even if they are not finalized yet.
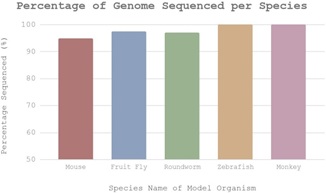 Figure 1: The chart depicts how much of the genome for each species has been annotated. The Zebrafish and Rhesus Monkey are both fully sequenced (100%). Note that the intervals begin at 50%.
Figure 1: The chart depicts how much of the genome for each species has been annotated. The Zebrafish and Rhesus Monkey are both fully sequenced (100%). Note that the intervals begin at 50%.
In another chart, the number of neurons per species was recorded (Figure 2). This allowed us to compare the complexity of the nervous system among the different species that were studied. The rhesus monkey had the most neurons out of the species studied (over a billion), which may indicate that it possesses more nervous complexity than a roundworm (less than a thousand neurons).
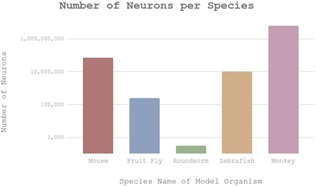
Figure 2: This bar graph compares the total number of neurons in the brain for each species used as a model organism. As a reference point, humans have 100 billion neurons in their brains.
The more closely related two species are, the more likely they are to share genetic material. To determine the evolutionary similarity between humans and each species, a cladogram was used to visualize the points at which each species diverged from the main lineage (Figure 3). The rhesus monkey, as expected, is strongly linked to humans, with mice following relatively close behind. Roundworms are very distantly related to humans, with our last common ancestor existing about 800 MYA.
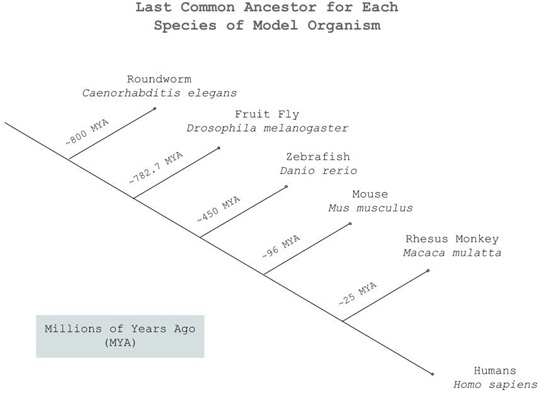 Figure 3: Cladogram depicting the points in evolutionary history at which each species of model organism diverges from humans.
Figure 3: Cladogram depicting the points in evolutionary history at which each species of model organism diverges from humans.
Genetic associations and phenotypic similarities
To identify the more specific attributes, the different genes that contribute to each disease were organized into a table with the disease phenotypes associated with all of the model organisms (Table 2). Starting with the number of genetic associations between each model organism in respect to each neurodegenerative disease, the column shows the number of genes reflected in the human genome that each model organism carries. This is also known as the number of genes that have been identified as affecting a particular disease. This information was also displayed in a graph (Figure 1). Mice have the most significant correlation to all three diseases with AD in particular. Zebrafish have the second highest amount of genes associated with AD, twice the number found in roundworms; however, they had very little correlation with PD and FTLD.
|
Diseases |
Organism |
Number of Genetic Associations / Disease Gene Homologs |
Phenotypes |
Benefits (B) / Limitations (L) |
References |
|
Alzheimer’s Disease |
Mouse |
15 |
Learning deficits |
B: The effects of the disease are clear to see - plaque buildup and damaged cells are easy to see. In addition, mice are cheap and easy to look after, and they are good test subjects for drugs L: Age dependent effects - may take a long time to observe effects in the mice. |
Cossins, Dan 2013 [11]
Lutz, Kathleen and Osborne, Melissa 2014 [12]
Hall, Alicia M. 2013 [13]
|
|
|
Fruit fly |
3 |
Eye degradation |
B: Relatively easy and inexpensive to culture, and there are very few ethical and regulatory restrictions on its use in laboratories. L: Fruit flies show disease phenotype during early stages of AD, whereas humans don’t show phenotypes until later stages. |
Clovis, Yoanne 2019 Part 1 [14,15]
Koon, Alex.C 2017 [16]
Tan, Florence Hui Ping [17]
|
|
|
Roundworm |
5 |
Progressive Paralysis |
B: Has simple nervous system that uses most of the neurotransmitters found in a mammalian brain. L: Roundworms have short lifespan - only 3.5 days naturally (without freezing). |
Clovis, Yoanne 2019 Part 1 & 2
Alexander, Adanna 2014 [18] |
|
|
Zebrafish |
10 |
Pathological Hyperphosphoryl ation |
B: Phenotypic effects of disease were clear - short tail, smaller head, and impaired brain development. L: Zebrafish brain structure and adult brain physiology needs to be better understood. |
Clovis, Yoanne 2019 Part 1 & 2
Newman, Morgan 2014 [19] |
|
Rhesus monkey |
N/A |
Neuron death and learning deficits |
B: Monkeys share many genetic similarities with humans. They reflect the entire biology of the disease that no other model organisms can. L: Raises many ethical concerns. There is little data on monkeys available for us. Little data is available for “disease genetic association” between rhesus monkeys and humans. |
Douglas, Grow 2014 [20]
Didier, E. S., et al. [21] |
|
|
Parkinson’s Disease |
Mouse |
9 |
Elevated dopamine release, mice are hyperactive |
B: Mice have longer life spans than that of roundworm and fruit fly. They also share more genetic similarities with humans L: According to studies, mice showed spontaneous locomotor activity, so they may be difficult to observe carefully. |
Cossins, Dan 2013
Lee, Yunjong 2012 [22] |
|
Fruit fly |
4 |
Reduced life span, male sterility |
B: Flies express proteins involved in Parkinson’s show cellular and behavioral phenotypes that resemble symptoms of human patients L: Short life span for studying Parkinson’s Disease. Not many effects can be observed during this time. |
Clovis, Yoanne 2019 Part 1
Koon, Alex. C 2017
Xiong, Yulan 2018 [23]
|
|
|
|
Roundworm |
4 |
Loss of dopaminergic neurons |
B: Good model for finding novel therapies. C. elegans have been shown to exhibit disruption and increased sensitivity to stress → similar to humans. L: Neuron connectivity differs from humans. Also, the molecular biology of dopamine and dopaminergic neurons is hard to study. |
Jason F. Cooper 2018 [24]
Clovis, Yoanne 2019 Part 2
Alexander, Adanna 2014 |
|
Zebrafish |
1 |
N/A |
N/A |
Lopes, Tomas 2013 [25] |
|
|
Rhesus monkey |
N/A |
N/A |
N/A |
N/A |
|
|
Frontotemporal Dementia |
Mouse |
7 |
Synaptic dysfunction and behavioral deficits |
B: The progressive motor neuron disease was visible; this test subject allows for easy visibility. L: They don’t accurately reflect responses to acute inflammatory stresses in humans. |
Cossins, Dan 2013
Lutz, Kathleen and Osborne, Melissa 2014
Roberson, Erik 2013 [26]
|
|
|
Fruit fly |
1 |
Reduced life span |
B: Fruit fly is a good model to study RNA toxicity involved in F. dementia. They have high offspring numbers and low cost for maintenance L: Fruit fly nervous system is very much different from the human and other mammalian nervous systems. |
Casci, Ian 2015 [27]
Koon, Alex. C 2017 |
|
|
Roundworm |
2 |
Uncoordinated movement |
B: Greater toxicity in worms expressing the mutant Tau protein are easily distinguishable L: Difficult to maintain, require a lot of effort |
Clovis, Yoanne 2019 Part 1
Alexander, A |
|
|
Zebrafish |
1 |
Accumulation of Tau throughout spinal cord, brain, and retina |
B: Red fluorescent cells can be seen expressing tau in embryos and behavioral deficits - easy visibility L: Its genome contains numerous duplicate genes, which means few mutant strains are available for studying neurodegenerative disease. |
Clovis, Yoanne 2019 Part 2
Solchenberger, Barbara 2015 [28]
Xi, Yanwei 2011 [29]
|
|
|
Rhesus monkey |
N/A |
N/A |
N/A |
N/A |
Table 2: The data in the table shows the results and observations of previous experiments conducted on the different species for each separate NDD. The specific genes that contribute to the diseases are also listed in the second column.
Fruit flies had genes ranging from one to four for the different diseases, and there was no recorded data with any of the diseases relating to the rhesus monkey.
Following genetic association is the disease phenotypes that each organism exhibit. Beginning with AD, different model organisms display different disease phenotypes. While some organisms display similar disease phenotypes to humans, others exhibit more distinct, easily observable phenotypes. For example, mice and rhesus monkeys with AD exhibit similar phenotypes as humans; they show learning and behavioral deficits. This similarity provides a more accurate reflection of human disease in animal models. On the other hand, fruit flies and roundworms with AD display different phenotypes such as eye degradation and progressive paralysis. Due to their physical nature, these phenotypes are easier to observe and study for researchers. In addition to disease phenotypes, a list of benefits and limitations were included. The list of benefits and limitations depict the strengths and weaknesses of each model organism in respect to the neurodegenerative disease in more detail. This information will be taken into consideration when comparing the model organisms in the discussion (Figure 4).
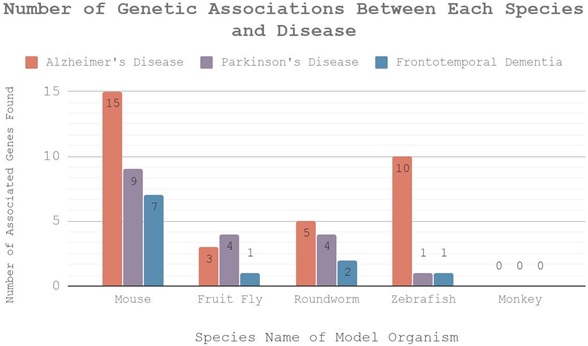 Figure 4: This graph illustrates the varying numbers of genes discovered for each model organism in the three diseases. The x-axis is the species name and the y-axis is the number of genetic associations found. The different colored bars represent different diseases: PD, AZ, and FTLD.
Figure 4: This graph illustrates the varying numbers of genes discovered for each model organism in the three diseases. The x-axis is the species name and the y-axis is the number of genetic associations found. The different colored bars represent different diseases: PD, AZ, and FTLD.
DISCUSSION
The usage of animal models plays a crucial role in the advancement of biomedicine and has been a cornerstone of medical advancement for many centuries. Mammals such as monkeys and mice have traditionally been the preferred models for biomedical research due to their close evolutionary relationship with humans. However, the use of mammals in research often raises ethical and budgetary concerns, such as the cost of larger mammal subjects and the ethics of using them. Due to these concerns, researchers have proposed to use non-mammalian model organisms such as roundworms, fruit flies and zebrafish. In the context of neurodegenerative diseases, although many model organisms have been used for research, no single species of model organism has been identified as most effective for research and development of treatment. In this research paper, we address this issue by providing genotypic and phenotypic comparisons of the advantages and limitations of many commonly used model organisms. Using this data and comparisons, we determine a single species of model organism that is most ideal for the general study and research of neurodegenerative diseases.
Table 1 depicts the life history traits of each organism. It showcases the organism’s generation time, when it reaches reproductive maturity, and how many children it can produce in one reproductive event. These particular traits were made into criteria because they are the three main characteristics that are considered when selecting an organism to model any sort of disease. Ideally, a model species should have a short generation time, reach sexual maturity in a short period of time, and have a large amount of offspring [30] These traits alone determine how usable and efficient an organism will be in a lab setting, no matter the topic of research it is involved in. A short generation time is necessary for a more efficient lab setting in which F1 and F2 generations are easily obtained. A substantial number of offspring allows for several different tests to be conducted on the progeny of a single pair. Reaching sexual maturity in a short period of time permits scientists to breed organisms in a timely manner. In Table 1, the rhesus monkey is observed to take the longest time to mature, and produces one child every 11 years under normal conditions. This indicates that monkeys are highly inefficient in a laboratory and may not be preferable for studies requiring several generations worth of experiments. On the other side of the spectrum, roundworms appear to have the most potential, as it has a brief generation time of 4 days, produces 300 offspring in a reproductive event, and only takes three days to sexually mature. The characteristics present in roundworms enable scientists to perform tests on multiple individuals after short periods of time.
In Figure 1, the chart depicts the progress on sequencing the genome of each individual species. While only the zebrafish and monkey have their DNA sequenced completely, all five organisms are shown to have at least 95% of their genome sequenced. Since the difference in percentages is relatively miniscule, it has little impact in deciding which model organism is ideal. Figure 2 displays the number of neurons per species. Here, roundworms are an outlier in the data, with very little neurons. Comparatively, rhesus monkeys come the closest to the actual amount of neurons in the human brain. This suggests greater complexity, which in turn has implications that mice and monkeys have elaborate nervous systems as opposed to roundworms. Considering that humans have the most neurons at approximately 100 billion, the more neurons an organism holds, it can be inferred that it becomes more similar to humans. The cladogram (Figure 3) visualizes the evolutionary lineage of all five species and when they diverge from humans. Rhesus monkeys are the most related to humans, having diverged 25 MYA. Because the more closely related two species are generally correlates with number of genetic associations overall, it can be assumed that rhesus monkeys share the most genetic material with humans, followed by mice.
Table 2 and Figure 4 consolidates data that include genotypic and phenotypic comparisons of different organisms for each neurodegenerative disease, and provides benefits and limitations of each model organism in the context of experimentation. One major goal of model organism research for human diseases is to identify genes that may harbor variation that affects the disease phenotypes, then verify if those genes will also have homologs in the human genome. Figure 4 reveals the number of genes that affect disease phenotypes for each NDD. For genes that cause Alzheimer’s Disease, Parkinson’s Disease, and Frontotemporal Dementia, mice share the greatest number of genes with humans for each disease (15, 9, and 7 respectively). Zebrafish have the second highest amount of genes associated with AD, twice the number found in roundworms; however, zebrafish had very little correlation with PD and FTLD. Fruit flies had genes ranging from one to four for the different diseases. For rhesus monkeys, although they possibly share more genes with humans than mice, there is no recorded data for genetic homologs of any of the diseases relating to the rhesus monkey due to ethical and budgetary concerns. These numbers show the extent of accuracy and resemblance to human diseases if researchers were to model those diseases on these organisms. Since mice share the greatest number of genes with humans, we can conclude that in terms of genetic association between model organisms and humans, mice would be the most ideal. In addition, according to Table 2, the exhibition of disease phenotypes show us the different symptoms model organism exhibit in respect to each neurodegenerative disease. In this criteria, the best model organism is determined in the context of which model organism display phenotypes that are most similar to those of human patients. In general, mice display phenotypes that are most similar to human patients across all three diseases. Although rhesus monkeys exhibit symptoms of neuron death and behavioral deficits in AD, which are found in human patients; the rhesus monkey doesn’t provide us with nearly as much data as mice. Fruit flies, roundworms, and zebrafish all display phenotypes that are easy to observe. However, since we determine the best model organism based on phenotypic similarities with human patients, they are not as ideal as mice to be used as a model organism because their phenotypes don’t align with human patients. Finally, Table 2 also provide a list of benefits and limitations for each model organism. These benefits and limitations were observed and concluded by researchers during previous experiments conducted on the different species for each separate neurodegenerative disease. To summarize the benefits and limitations of each model organism, in general non-mammalian model are more cost effective, display symptoms that are easier to observe, and they don’t raise ethical concerns. Nonetheless, limitations of non-mammalian models outweigh their benefits. Since non-mammalian models have short life span, they typically display phenotypes during early stages of the disease. However, human patients with NDDs don’t display phenotypes until later stages of the disease. This difference makes mammalian model organisms like mice a better and more accurate model than non-mammalian model organisms such as roundworms, zebrafish, and fruit flies. However, this doesn’t mean non-mammalian organisms are not beneficial at all during the research process. Non-mammalian model organisms are typically used in early research to deliver fast answers to a discovery problem - such as the function of a gene, or to define novel therapeutic entry points. Since the way we determine the best model organism is based on their genetic and phenotypic similarities of human patients, mice are considered to be the most ideal model organism for the general study and research of neurodegenerative diseases.
In our research, we realize that there are caveats to our findings. One would be the ethics of non-human model organisms. Animal rights organizations such as PETA are actively campaigning against animals being used in research, and the US government has set ethical standards in using animals for modeling human diseases. Since the research also involves NDDs, there have been concerns about the effects that these could show to mammalian nervous systems, as they are similar to human nervous systems. Thus, data on larger mammals, such as the rhesus monkey, was not readily available to us.
However, we were able to find data on the other non-mammalian model organisms. Another caveat of our research could have been bias towards certain organisms. Due to the nature of a meta-analysis, the data we collected could have held a potential bias towards a certain model organism. For example, a scientist conducting research could potentially choose one organism over another due to personal reasons. Additionally, a scientist may have potentially miscounted or not observed a phenotype that a model organism showed. Thus, we may not know a specific phenotype displayed by a model organism due to a scientist not fully recording their research.
The ultimate goal of our research was to find an ideal model organism to help study NDDs. We initially hypothesized that the rhesus monkey would be the ideal model organism due to its genetic similarity to humans and its complex nervous system. However, this hypothesis was rejected as we combed through our data. When we hypothesized that the rhesus monkey was the ideal model organism, we did not take into account the ethics behind choosing non-primate models. We learned, however, that this was the reason why there was not sufficient data to support our hypothesis. Many members of the public are opposed to researching upon primates. Instead, we found that the ideal model organism is the mouse (Mus musculus). This is due to many reasons, including the genetic associations with NDDs in humans, as well as being a mammal and having a more complex nervous system. Mice also have many applications in biotechnology; for example, many of the genetic associations found using mice were discovered through transgenesis. This method allows for a more direct understanding of the correlation between certain human genes and the phenotypic effects of each NDD studied. We observed that most of the transgenic model organisms were of the species mus musculus, and because of this it can be concluded that the utilization of transgenesis is more efficient on mice. The mouse is also considered an ethical option that the US government is compliant with using. According to our research, the data we have collected and the conclusions we have come to mean that mice should be used to study NDDs in the future as they show similar genotypic and phenotypic characteristics to humans when introduced to NDDs.
This research aimed to standardize the selection of a model organism for future research. By finding similarities in the NDDs studied, we were able to compare model organisms in their phenotypic responses to the diseases. Standard units of measurement are deeply embedded in scientific academia; it is only logical that there should be standard organisms for NDDs, especially since many of them share similar traits. We hope that our comparisons will enable future scientists to effectively choose model organisms for their research.
REFERENCES
- https://www.neurodegenerationresearch.eu/what/
- Rohn TT, Kim N, Isho FN, Mack JM (2018) The Potential of CRISPR/Cas9 Gene Editing as a Treatment Strategy for Alzheimer’s Disease. Journal of Alzheimer's disease & Parkinsonism 8: 439.
- Klein C, Westenberger A (2019) Genetics of Parkinson’s Disease. Cold Spring Harb Perspect Med 2: a008888.
- Cannon JR, Greenamyre JT (2011) The role of environmental exposures in neurodegeneration and neurodegenerative diseases. Toxicological Sciences 124: 225-250.
- Gatto NM, Cockburn M, Bronstein J, Manthripragada AD, Ritz B (2009) Well-water consumption and Parkinson’s disease in rural California. Environmental health perspectives 117: 1912-1918.
- Lallani SB, Villalba RM, Chen Y, Smith Y, Chan AWS (2019) Striatal Interneurons in transgenic Nonhuman primate Model of Huntington’s Disease. Scientific Reports 9: 3528.
- Habermann B, Hines D, Davis DHL (2013) Caring for parents with neurodegenerative disease: A qualitative description. Clinical nurse specialist CNS 27: 182-187.
- Findley LJ (2007) The economic impact of Parkinson's disease. Parkinsonism & Related Disorders 13: S8-S12.
- Rosa MAG, Gasperi RD, Elder GA (2012) Modeling human neurodegenerative diseases in transgenic systems. Human Genetics 131: 535-563.
- Hammond TR, Marsh SE, Stevens B (2019) Immune Signaling in Neurodegeneration. Immunity 50: 955-974.
- https://www.the-scientist.com/do-mice-make-bad-models-39790.
- Lutz CM, Osborne MA (2014) Optimizing Mouse Models of Neurodegenerative Disorders. Medscape 9: 67-75.
- Hall AM, Roberson ED (2013) Mouse models of Alzheimer's disease. Brain Research Bulletin 88: 3-12.
- https://invivobiosystems.com//disease/worms-flies-fish-comparison-common-model-organisms/
- https://nemametrix.com/disease/worms-flies-or-fish-a-comparison-of-common-model-organisms-part-2-models-for human-diseases/
- Koon AC, Chan HYE (2017) Drosophila Melanogaster As a Model Organism to Study RNA Toxicity of Repeat Expansion-Associated Neurodegenerative and Neuromuscular Diseases. Frontiers in Cellular Neuroscience 11: 70.
- Tan FHP, Azzam G (2017) Drosophila Melanogaster: Deciphering Alzheimer's Disease. The Malaysian Journal of Medical Sciences 24: 6-20.
- Alexander AG, Marfil A, Li C (2014) Use of Caenorhabditis Elegans as a Model to Study Alzheimer's Disease and Other Neurodegenerative Diseases. Front Genet 5: 279.
- Newman M, Ebrahimie E, Lardelli M (2014) Using the Zebrafish Model for Alzheimer's Disease Research. Frontiers in Genetics 5: 189.
- Grow DA, McCarrey JR, Navara CS (2016) Advantages of Nonhuman Primates as Preclinical Models for Evaluating Stem Cell-Based Therapies for Parkinson's Disease. Stem Cell Research 17: 352-366.
- Didier ES, Maclean AG, Mohan M, Didier PJ, Lackner AA, et al. (2016) Contributions of Nonhuman Primates to Research on Aging. Vet Pathol 53: 277-290.
- Lee Y, Dawson VL, Dawson TM (2012) Animal Models of Parkinson's Disease: Vertebrate Genetics. Cold Spring Harb Perspect Med 2: a009324.
- Xiong Y, Yu J (2018) Modeling Parkinson's Disease in Drosophila: What Have We Learned for Dominant Traits? Front Neurol 9: 228.
- Hall AM, Roberson ED (2013) Mouse models of Alzheimer's disease. Brain research bulletin 88: 3-12.
- Fonseca TL, Correia A, Hasselaar W, Linde HC, Wilemsen R, et al. (2013) The Zebrafish Homologue of Parkinson's Disease ATP13A2 Is Essential for Embryonic Survival. Brain Research Bulletin 90: 118-126.
- Roberson ED (2012) Mouse Models of Frontotemporal Dementia. Annals of Neurology 72: 837-849.
- Casci I, Pandey UB (2015) A Fruitful Endeavor: Modeling ALS in the Fruit Fly. Brain Research 1607: 47-74.
- Solchenberger B, Russell C, Kremmer E, Haas C, Schimid B (2015) Granulin knock out zebrafish lack frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis pathology. PLoS One 10: e0118956.
- Xi Y, Noble S, Ekker M (2011) Modeling neurodegeneration in zebrafish. Current Neurology and Neuroscience Reports 11: 274-282.
- Hmelja K, Justice M (2019) From gene to treatment: supporting rare disease translational research through model systems. Dis Model Mech 12: dmm039271.
Citation: Zhang XE (2020) The Potential of Distinct Species of Non-Human Model Organisms for Research of Neurodegenerative Diseases. J Alzheimer’s Neurodegener Dis 6: 047.
Copyright: © 2020 Xiucheng Eric Zhang, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

