
The Revolutionizing Prospects of Next Generation DNA Sequencing of Microorganisms for Microbiological and AMR Identification and Characterization-the GMI Singapore Sequencing Statement
*Corresponding Author(s):
Joergen SchlundtSchool Of Chemical And Biomedical Engineering, Nanyang Technological University (NTU), 62 Nanyang Drive, Singapore
Tel:+65 98351808,
Email:jschlundt@ntu.edu.sg
Abstract
A cascade of technological DNA sequencing advancements has decreased the cost of Whole Genome Sequencing (WGS) very fast. At the same time, the enormous potential of WGS in the surveillance of microorganisms and Antimicrobial Resistance (AMR) is increasingly realized. One Health Solutions-based on cross-sectoral typing and source attribution are increasingly spreading in relation to foodborne disease and AMR prevention. Collaboration for disease prevention between human, veterinary and food sectors is already instituted in some countries and has in some cases resulted in significant reduction in foodborne disease risk, as well as AMR risk. The success of One Health risk mitigation is to a large part based on the potential to link relevant microbiological data from different sectors (animal, food and human) as the basis for science-based decision support to reduce the risk of foodborne diseases and AMR. Such linkage is only possible if the methodologies used, or at least the outcome microbiological characterizations, are comparable. While this has been a major problem in the past, novel DNA sequencing methodologies are now providing promising new potential. The case is made for the potential to develop a global, equitable system of whole microbial genome databases to aggregate, share, mine and use microbiological genomic data, to address global food safety and public health challenges and most importantly to reduce foodborne and infectious disease burden.
Keywords
Antimicrobial resistance; Food safety; Genomic data sharing; Global microbial identifier; Microbiology; Next generation sequencing; One health; Surveillance; Zoonotic diseases
INTRODUCTION
The disease burden of zoonotic diseases has been generally recognized, but not very well characterized until recently. While significant zoonotic diseases (e.g. Rabies, Lyme’s disease) relate to the direct contacts with animals (bites), most important zoonoses relate in some way to animals in the food production chain. For many decades, food safety events in most countries have been significantly affected by a lack of collaboration between the animal health, the food control and the human health sectors. Nevertheless, it was primarily the outbreaks of SARS (Severe Acute Respiratory Syndrome), zoonotic influenza (avian flu-H5N1 and swine flu-H1N1) and BSE (Bovine Spongiform Encephalopathy) which alerted the world to the need for a One Health approach. This is a bit disappointing since we have known for some time that endemic cases of traditional foodborne zoonoses (salmonellosis, campylobacteriosis, listeriosis, norovirus) causes a much larger-and continuous-disease burden than these dramatic global outbreaks [1].
In the beginning of this millennium, zoonotic influenza outbreaks spread very quickly, either in the Animal Population (e.g. H5N1) or directly in the Human Population (e.g. H1N1) and formed a global threat for human health. H1N1 was therefore characterized by the World Health Organization (WHO) as a pandemic. Although, as mentioned, the human disease burden related to the endemic bacterial zoonoses is many fold higher than these influenza outbreaks, it is basically these relatively few but fast spreading outbreaks that have put One Health on the global agenda. The failure to predict, monitor and control the spread of these diseases in animals presented regulators and especially politicians with a wake-up call and made them demand (better) cross-sectoral collaboration between the animal and human health sectors.
Therefore, the one health concept was formulated to characterize the benefit of cross-sectoral collaboration. Taking the cue from high-level intergovernmental meetings, the relevant intergovernmental organizations suggested the set-up of systems and solutions based on one health solutions. The first such intergovernmental description of the one health concept is from the publication “Contributing to One World, One Health” [2], but the concept was later more clearly enunciated as “One Health” in the seminal paper by WHO,
World Organisation for Animal Health (OIE) and Food and Agriculture Organization of the United Nations (FAO), “The FAO-OIE-WHO Collaboration: Tripartite Concept Note-Sharing responsibilities and coordinating global activities to address health risks at the animal-human-ecosystems interfaces” [3]. It can only be characterized as astounding that these three intergovernmental organizations had not at an earlier stage clearly described this need for coordination, although it must be recognized that in the food safety standards area the FAO/WHO Codex Alimentarius Commission as well as the FAO/WHO Expert Committees have actually been a beacon for intersectoral coordination since at least 1963!
Collaboration between sectors is generally a benefit in many respects, especially the potential linkage of relevant microbiological data is important. Since there is an obvious need for science-based decision support to reduce the risk of zoonotic diseases, the specific comparison of microbiological isolates from animal, food and human is now recognized as crucial. Such comparison is only possible if the methodologies used, or at least the outcome microbiological characterizations, are comparable. While this has been a major problem in the past, novel DNA sequencing methodologies are now providing promising new potential (see Section New characterization and typing methodology).
One of the fastest growing problems in the One Health arena is AMR in zoonotic and other microorganisms, which is an emerging public health issue all over the world. This problem will have societal and economic impact in developing as well as developed countries, spanning all One Health sectors from the environment, over husbandry animals to food and finally human.
A One Health approach to AMR may help bridge gaps in the levels of commitment being shown to each sector and enable policy development that is inclusive, sensitive and sufficiently flexible to accommodate the varying needs of different sectors, countries and regions. Over the last 20 years a number of publications have documented a global trend of a rising AMR threat [4-7], notably including emergence of resistance against antimicrobials that are considered critically important in human medicine and in Multidrug Resistant (MDR) infections [8,9]. Some of the recent serious outbreaks of Antibiotic Resistant (AMR) foodborne disease underlines this trend (e.g. the EHEC (Enterohaemorrhagic Escherichia coli) outbreak in Germany [10], but it is-again-important to note that although outbreaks are often picked up by the press, the real AMR problem is actually an endemic problem, not an epidemic problem [11]. In fact, it is estimated that in the absence of significant risk mitigation, the number of annual deaths globally from AMR will reach 10 million, significantly surpassing the present 7-8 million deaths from cancer [12].
The use of antimicrobials in humans and animals has resulted in a selective pressure for AMR microorganisms, contributing significantly to the human health problem of AMR bacteria. A WHO publication [13], covers the broader scope of AMR in relation to both animals and humans. Thus, a One Health approach has explicitly been proposed by international organizations to mitigate the risk of AMR. In particular, the use in animals as Antimicrobial Growth Promoters (AGP) is questionable, as the concentrations used are sub-therapeutic, thus resulting in selection for resistance but not efficient kill of relevant microorganisms. One Health systems, including cross-sectoral typing, may provide scientific basis for risk mitigation relative to microbiological and AMR problems in both human and animal reservoirs (Figures 1 and 2). However, there seems to be a problem in assessing the relative importance of human and animal use and the veterinary and medical professions are still hotly debating this [14-17]. It is therefore important to be able to compare data on AMR from both the animal and the human side enabling integrated source attributions. Again, novel DNA sequencing methodologies are now providing promising new potential (see Section New characterization and typing methodology).
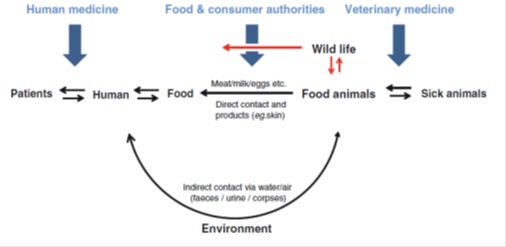 Figure 1: Schematic presentation of important microbial transmission routes via which the human and (food) animals are in contact with each other. In blue control mechanisms are shown and in red some of the transmission routes that are more difficult to control. Via the environment transmission may take place of microorganisms present in excretion products and in diseased animals and people. In addition, wildlife constitutes a risk, as it holds a broad spectrum of diseases, including many highly pathogenic diseases.
Figure 1: Schematic presentation of important microbial transmission routes via which the human and (food) animals are in contact with each other. In blue control mechanisms are shown and in red some of the transmission routes that are more difficult to control. Via the environment transmission may take place of microorganisms present in excretion products and in diseased animals and people. In addition, wildlife constitutes a risk, as it holds a broad spectrum of diseases, including many highly pathogenic diseases.
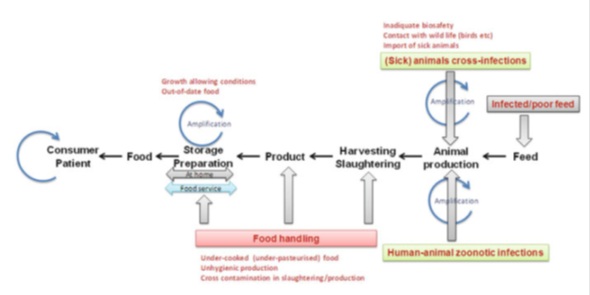 Figure 2: Farm-to-fork scheme showing how infectious diseases may travel through the food chain. Along the chain we have indicated some of the most prevalent causes of infectious disease introduction.
Figure 2: Farm-to-fork scheme showing how infectious diseases may travel through the food chain. Along the chain we have indicated some of the most prevalent causes of infectious disease introduction.
SOLUTIONS BASED ON MICROBIOLOGICAL SURVEILLANCE
One Health Solutions-based on cross-sectoral typing and source attribution are increasingly spreading in relation to zoonotic disease and AMR prevention. Building on the idea of One Health to guide disease prevention collaboration between human, veterinary and food sectors has already been instituted in some countries and has in some cases resulted in significant reduction in foodborne disease risk, as well as AMR risk.
In Denmark, a program to reduce/eliminate Salmonella in egg-laying hens started with a simple and inexpensive serological surveillance of egg laying hen flocks, followed by a program of surveillance and eradication of infected broiler flocks. The effect of the program was evaluated through Salmonella typing linking animal isolates to cases of human salmonellosis, through using comparable detection and sero- and geno-typing techniques, which enabled comparison and sharing of data, thus clearly following One Health principles. The program resulted in a very significant reduction in human salmonellosis and in the end in the total elimination of Salmonella in egg-laying hens in Denmark [18].
Many international organizations have generated guidelines to disseminate information aboutfood related zoonoses, such as for instance WHO's Global Foodborne Infections Network (GFN) (www.who.int/gfn), WHO/FAO INFOSAN (International Food Safety Authorities Network) (https://www.who.int/activities/responding-to-food-safety-emergencies-infosan), the European Food Safety Authority (EFSA) (www.efsa.europa.eu/en/topics/topic/zoonoticdiseases ), Food net from the US Centres for Disease Control and Prevention (www.cdc.gov/foodnet ) and others. The goals of these networks are essentially the same: To promote integrated, laboratory based surveillance and intersectional collaboration among human health, veterinary and food-related disciplines to reduce the risk of foodborne infections.
One Health approaches to mitigate zoonotic foodborne diseases, as well as AMR are important at international and national level. However, most of this work is presently done at international level, i.e. through the work of Expert Groups under WHO, OIE and FAO, or in the EU system through EFSA Committees. An example of the early application of a One Health approach relative to national AMR surveillance and risk mitigation is the Danish program to contain AMR zoonoses, DANMAP [19-21]. Starting in 1995, the Danish government decided to set up the Danish Integrated Antimicrobial Resistance Monitoring and Research Program (DANMAP). The objectives of DANMAP are:
- To quantitatively monitor the consumption of antimicrobials used in (food) animals and humans;
- To quantitatively monitor the occurrence of AMR in (zoonotic) bacteria in animals, food and humans;
- To identify routes of transmission and areas for further research.
Data from the DANMAP programme was used as science-based decision support, ultimately resulting in the banning of AGP use in food animals in Denmark and later (2006) in all of EU.
Notwithstanding such national examples of science-based action and good outcome, a stronger evidence base to inform effective policy interventions across both the human and animal sectors, is needed. Dar et al. [22], suggests a need for the sharing of experience covering three domains:
- Responsible antimicrobial use;
- Surveillance and;
- Infection prevention and control, considering which policies are likely to be most effective at national and regional levels.
In general, it must now be accepted that integrated surveillance across humans and animals is both possible and can lead to effective risk reduction. However, the traditional typing systems are cumbersome, time-consuming and in many cases not generally applicable for instance for source attribution (source attribution using traditional microbiological typing systems are effective only used for Salmonella). Thus, again, the need for better typing tools is clear.
NEW CHARACTERIZATION AND TYPING METHODOLOGY
The technological advances and decreasing costs of DNA sequencing now provide tools to investigate the dynamics of infection-even in real time-through the analysis of microbial genome diversity. The projected significant increase in whole (microbial) genome sequences will likely also enable a much better understanding of the pathogenesis of the infection and the molecular basis of the host response to infection.
Data in this area should be comparable between labs and indeed between countries, preferably in open-source systems. There is therefore an obvious need to develop a global system of microbial DNA databases to aggregate, share, mine and use microbiological genomic data, to address global public health and clinical challenges and most importantly to identify and diagnose infectious diseases [23] (see Section The need for a global system for DNA sharing and use).
Importantly, in the future, rapidly developing technologies such as the use of novel, continuously developing, DNA sequencing methods for microbiological identification and characterization will enable national and global surveillance of both communicable diseases and AMR in humans, animals and food in fully integrated systems [24].
Nearly all of the most important human pathogens are either zoonotic or originated as zoonoses [25]. Striking examples include HIV/AIDS and Spanish influenza, which started by interspecies transmission of the causative agents [26] and have caused millions of deaths worldwide and more recently SARS and MERS (Middle East Respiratory Syndrome) coronaviruses and H1N1 and H5N1 influenza A viruses. Realizing that detection and surveillance form the backbone of all systems currently used to control infectious diseases worldwide and that disease (including foodborne disease) surveillance is still typically targeted at a relatively limited number of specified diseases, a more effective and rational approach to the prevention of microbial threats is essential [26]. And this rational approach must include some sort of assurance of methodological linkage between isolates from humans, animals as well as food and the environment. With recent technological advances and declining costs in the Next Generation Sequencing (NGS) field, these tools will play an increasingly important role in the surveillance of existing pathogens, as well as identification of new and previously unrecognized pathogens in both animals and humans as well as food and environment. Thus, a very significant increase in microbial DNA and RNA sequences, including Whole Genome Sequences (WGS) is to be expected.
Surveillance is done globally to monitor trends in endemic diseases (e.g. influenza, dengue and salmonellosis), to monitor eradication efforts (polio, measles, brucellosis), or to signal unusual disease activities. Previous (‘traditional’) molecular diagnostic tools, which rely on the recognition of short pieces of unique genome sequence (e.g. PCR and microarray (biochip) technologies) have been used routinely for some time in clinical diagnostic and surveillance settings. Although the partial DNA information can be sufficient for patient management and basic surveillance objectives, from a public health perspective, this trend is worrisome since recombination of viral genomes may generate future viral threats (e.g. influenza A viruses are able to undergo reassortment if a single cell is concurrently infected with more than one virus). These reassortment events can dramatically change the evolution of influenza A viruses in a certain host and lead to new epidemics and pandemics; and such events may easily be missed when/if surveillance is relying on molecular diagnostic tools that target small microbial genome fragments.
WGS determines the complete genome sequence of an organism, not only fragments, which can have important implications. During the recent outbreak of MERS coronavirus in the Middle East, analysis of small genome fragments did not provide sufficient phylogenetic signal for reliable typing of virus variants [27]. Classically, whole microbial genome sequences were determined by PCR and Sanger sequencing. Nowadays, next NGS techniques are used increasingly to genotype microorganisms in almost any microbial setting [28].
THE NEED FOR A GLOBAL SYSTEM FOR DNA SHARING AND USE
The Global Microbial Identifier (GMI-www.globalmicrobialidentifier.org) initiative is a not-for-profit international consortium comprising scientists from over 55 countries suggesting international agreements about the sharing of DNA data through the creation of open, accessible global databases.
The GMI initiative, aims to build a database of whole microbial genome sequencing data linked to relevant metadata, which can be used to identify microorganisms, their communities and the diseases they cause (Table 1). Such interactive database (as opposed to existing passive databases) would contain full DNA or RNA data from microorganisms (bacteria, virus, fungi and parasites) for the immediate identification and characterization of isolates, for the identification of relevant genes and for the comparison of genomes to detect outbreaks and emerging pathogens. Importantly, this potential future database or system of databases will not only cover pathogens, but indeed all microbiological species [29].
|
Recognizing that risks related to the spread of dangerous microorganisms in humans, plants, food, animals and environment, compounded by the growing threat of antimicrobial resistance (AMR), are a global concern. |
|
• Recent Public health and One Health initiatives attempts to protect citizens from health risks posed by pathogenic microorganisms, which could cause as many as 18-19 million deaths annually including 10 million due to AMR by 2050 (more than present deaths from cancer); • Novel developments in sequencing DNA from microorganisms is revolutionizing the detection and prevention of the spread of such microorganisms and AMR. Equal access and implementation of such new sequencing technology between countries can dramatically reduce the global burden of disease by enabling a novel, real-time surveillance of all animal and human diseases and food safety risks. • Global sharing of sequencing results will allow for the early detection of emerging threat and rapid identification, investigation and prevention of national, regional and global disease outbreaks. |
|
Scientists, gathered at the 12th Meeting of the GMI (Global Microbial Identifier) in Singapore, urge all countries to consider the public, animal and plant health, food safety and economic benefits of introducing a global mechanism for the sharing and analysis of DNA sequences |
|
• The GMI initiative is a not-for-profit international consortium comprising scientists from over 55 countries collaborating and sharing sequencing data for microorganisms, enabling efficient global surveillance and a new understanding of the importance of microorganisms in general. Membership in GMI is entirely open and encouraged for everyone working in this field. • GMI provides a framework for coordinating DNA data collection and analyses of microorganisms with the goal of open sharing of sequence data. GMI provides validation guidance for both the sequencing data collection and analyses, as well as capacity building efforts for developing countries • In a fully realized global sequencing database (or interconnected databases), microorganisms can be rapidly characterized in context of their global diversity, controlling disease outbreaks, enacting food recalls, providing a resource for preventive controls and tracking the spread of AMR. • The use of sequencing methodologies revolutionizes our understanding and management of plant, animal, environmental, human health and food safety. Optimal use is dependent on policies and the willingness and ability of countries to share genomic sequences across borders and in real-time. |
|
Government and intergovernmental organizations must implement sequencing data sharing policies and mechanisms, ensuring equitable access and benefits to people worldwide, with the vision to improve global human health[1]. |
Table 1: Singapore Sequencing Statement, agreed at the GMI 12 meeting in Singapore, June 2019.
To harness the full potential of novel DNA sequencing, a shared global database of DNA, RNA and genomes linked to relevant metadata and the necessary software tools need to be generated, which is the key focus of the GMI initiative. This tool will ideally be used in amongst others in the diagnosis of infectious diseases in humans and animals, in the identification of microorganisms-including beneficial microorganisms and microorganisms for technological use - in food and environment.
A cascade of technological NGS advancements has decreased the cost of WGS very fast. At the same time, the enormous potential of WGS in the surveillance of infectious diseases and AMR [30], has been demonstrated in many studies now, including the tracking and tracing of the cholera outbreak in Haiti in 2010 [31], the EHEC outbreak in Germany in 2011 [32] and others. During the EHEC outbreak, scientists from around the globe performed NGS and shared their results for analysis. Similar collaborations exist globally during emerging viral infections such as MERS coronavirus and in recent Wuhan pneumonia outbreak that is caused by a novel coronavirus [33,34]. Within a month after the first case was reported, the “initial’ sequence of the novel coronavirus was released publicly by the consortium [35] and the coronavirus research community begins conducting research in their area of expertise [34]. This highlights the importance of sharing WGS data between scientists and its impact on global public health. The sharing of WGS can be used to compare the DNA sequence of an isolate to standardized reference sequences, thereby enabling investigation/documentation of important phenotypic characteristics of an organism, such as virulence factors or AMR. WGS has the highest possible resolution for isolate sub typing and is therefore optimal for rapid tracing and outbreak investigation. Comprehensive genome databases linked to standardized epidemiological data would be an invaluable resource for animal health, food safety and public health, as well as for research in general, including research into non-pathogenic microorganisms, which are obviously important in food and environmental, and even in clinical microbiology. It is, however, important to ensure that WGS surveillance data are of sufficient quality to inform decision-making. This also means that there is an urgent need to define internationally standardized formats for metadata.
The need to integrate DNA databases and to harmonize and maybe globalize, data collection and sharing has been recognized by the scientific community for some time [36]. Further integration of these databases and linking the genomic data to metadata for optimal prevention of infectious diseases and to make it fit for other uses including routine diagnostics, is a new challenge.
While the NGS revolution has now been introduced in most rich countries, this major change provides an even more interesting potential for developing countries, creating a potential for a diagnostic leap-frog in these countries. NGS holds the potential of a simple one-size-fits-all tool for diagnosis of all infectious diseases, thereby dramatically improving public health in developing countries. At a systemic level, the use of NGS will enable uniform laboratory-, reporting- and surveillance-systems not only relative to human health, but reaching out to the identification of microorganisms in all other habitats, including animals, plants and the environment: A true One Health approach.
Recent studies have shown that it is possible to determine the species, type as well as the antimicrobial/antiviral susceptibility of both bacterial and viral pathogens, even when using sequencing directly on clinical samples [30]. This would be even more valuable for clinical laboratories in developing countries that do not currently have the same diagnostic capacities as most developed countries [37].
As NGS technology spreads more globally, there is an obvious potential to develop a global system of whole microbial genome databases to aggregate, share, mine and use microbiological genomic data, to address global public health and clinical challenges and most importantly to identify and diagnose infectious diseases.
If created, a global DNA database system should be deployed in a manner, which promotes equity in access and use of the current technology. If the system is set up in an ‘open access’ format, it would likely enable comprehensive utility of NGS in developing countries, since open databases and relevant open algorithm platforms could be used for immediate translation of sequence data to microbial identity and antimicrobial resistance pattern [37]. In general, it is necessary to have a comprehensive database of all known DNA sequences to make full use of local derived DNA sequence to characterize microbial isolates and track epidemics.
[1]Many countries are currently re-thinking laws and policies that address the management and conservation of biodiversity, as well as the protection of the public’s health and the promotion of Open Science. GMI urges governments and intergovernmental organizations to use this window of opportunity to support and regulate global microbial DNA sequence data sharing.
ABBREVIATIONS
|
Antimicrobial Growth Promoters |
AGP |
|
Antimicrobial Resistance |
AMR |
|
Bovine Spongiform Encephalopathy |
BSE |
|
Enterohaemorrhagic Escherichia Coli |
EHEC |
|
European Food Safety Authority |
EFSA |
|
Food and Agriculture Organization of the United Nations |
FAO |
|
Global Foodborne Infections Network |
GFN |
|
Global Microbial Identifier |
GMI |
|
International Food Safety Authorities Network |
INFOSAN |
|
Middle East Respiratory Syndrome |
MERS |
|
Multidrug Resistant |
MDR |
|
Next Generation Sequencing |
NGS |
|
Severe Acute Respiratory Syndrome |
SARS |
|
Whole Genome Sequencing |
WGS |
|
World Health Organization |
WHO |
|
World Organisation for Animal Health |
OIE |
DECLARATIONS
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of supporting data
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Funding
This study was supported by funding from Nanyang Technological University (NTU) Research Initiative.
Authors' contributions
J.S. was a major contributor to article writing and revision. M.Y.F.T. contributed to article writing and revision.
Acknowledgement
Not applicable.
Authors' information
Joergen Schlundt
ORCID: https://orcid.org/0000-0002-3336-2935
Moon Yue Feng Tay
REFERENCES
- Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, et al. (2015) World Health Organization Global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 12: 1001923.
- FAO Animal Production and Health Division, World Health Organization, World Organisation for Animal Health, UNICEF, UN System Influenza Coordination, World Bank (2008) Contributing to one world, one health. A strategic framework for reducing risks of infectious diseases at the animal-human-ecosystems interface.
- FAO-OIE-WHO Collaboration (2010) Sharing responsibilities and coordinating global activities to address health risks at the animal-human-ecosystems interfaces: A Tripartite Concept Note.
- World Health Organization (2001) WHO global strategy for containment of antimicrobial resistance. World Health Organization, Geneva, Switzerland.
- DANMAP (2018) The Danish integrated antimicrobial resistance monitoring and research programme.
- European Centre for Disease Prevention Control (2010) Methodology protocol for estimating burden of communicable diseases. ECDC Stockholm.
- Aarestrup F (2012) Sustainable farming: Get pigs off antibiotics. Nature 486: 465-466.
- Potron A, Kalpoe J, Poirel L, Nordmann P (2011) European dissemination of a single OXA-48-producing Klebsiella pneumoniae clone. Clin Microbiol Infect 17: 24-26.
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, et al. (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis 10: 597-602.
- Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, et al. (2011) Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104: H4 outbreak by rapid next generation sequencing technology. PLoS One 6: 22751.
- MacIntyre CR, Bui CM (2017) Pandemics, public health emergencies and antimicrobial resistance- putting the threat in an epidemiologic and risk analysis context. Arch Public Health 75: 54.
- O’Neill J, Davies S, Rex J, White L, Murray R (2016) Review on antimicrobial resistance, tackling drug-resistant infections globally: Final report and recommendations. London, UK.
- WHO (2011) Tackling antibiotic resistance from a food safety perspective in Europe. World Health Organization, Geneva, Switzerland.
- Phillips I, Casewell M, Cox T, De Groot B, Friis C, et al. (2004) Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J Antimicrob Chemother 53: 28-52.
- Karp BE, Engberg J (2004) Comment on: Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J Antimicrob Chemother 54: 273-278.
- Smith DL, Dushoff J, Morris JG (2005) Agricultural antibiotics and human health. PLoS Med 2: 232.
- Price LB, Stegger M, Hasman H, Aziz M, Larsen J, et al. (2013) Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 3: 00520.
- Wegener HC, Hald T, Lo Fo Wong D, Madsen M, et al. (2003) Salmonella control programs in Denmark. Emerg Infect Dis 9: 774-780.
- WHO (2003) Impacts of antimicrobial growth promoter termination in Denmark: The WHO international review panel's evaluation of the termination of the use of antimicrobial growth promoters in Denmark: Foulum, Denmark 6-9 November 2002. World Health Organization, Geneva,
- Hammerum AM, Heuer OE, Emborg H-D, Bagger-Skjøt L, Jensen VF, et al. (2007) Danish integrated antimicrobial resistance monitoring and research program. Emerg Infect Dis 13: 1633-1639.
- Aarestrup FM, Jensen VF, Emborg H-D, Jacobsen E, Wegener HC (2010) Changes in the use of antimicrobials and the effects on productivity of swine farms in Denmark. Am J Vet Res 71: 726-733.
- Dar OA, Hasan R, Schlundt J, Harbarth S, Caleo G, et al. (2016) Exploring the evidence base for national and regional policy interventions to combat resistance. Lancet 387: 285-295.
- Allard MW, Strain E, Melka D, Bunning K, Musser SM, et al. (2016) Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 54: 1975-1983.
- Aarestrup FM, Brown EW, Detter C, Gerner-Smidt P, Gilmour MW, et al. (2012) Integrating genome-based informatics to modernize global disease monitoring, information sharing, and response. Emerg Infect Dis 18: 1.
- Taylor LH, Latham SM, Woolhouse ME (2001) Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356: 983-989.
- Osterhaus A, Smits S (2012) Genomic and personalized medicine (2nd ed.). Elsevier, Amsterdam, the Netherlands.
- Smits SL, Raj VS, Pas SD, Reusken CB, Mohran K, et al. (2015) Reliable typing of MERS-CoV variants with a small genome fragment. J Clin Virol 64: 83-87.
- Smits SL, Osterhaus AD (2013) Virus discovery: One step beyond. Curr Opin Virol 3: 1-6.
- Wielinga PR, Hendriksen RS, Aarestrup FM, Lund O, Smits SL, et al. (2017) Global microbial identifier. Applied Genomics of Foodborne Pathogens, Springer.
- Hasman H, Saputra D, Sicheritz-Ponten T, Lund O, Svendsen CA, et al. (2014) Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J Clin Microbiol 52: 139-146.
- Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, et al. (2011) Population genetics of Vibrio cholerae from Nepal in 2010: Evidence on the origin of the Haitian outbreak. mBio 2: 00157-11.
- Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, et al. (2011) Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104: H4 outbreak by rapid next generation sequencing technology. PloS One 6: 22751.
- Normile D (2020) Mystery virus found in Wuhan resembles bat viruses but not SARS, Chinese scientist says. American Association for the Advancement of Science.
- Cohen J (2020) Chinese researchers reveal draft genome of virus implicated in Wuhan pneumonia outbreak. American Association for the Advancement of Science.
- Holmes EC (2020) Initial genome release of novel coronavirus.
- Aarestrup FM, Brown EW, Detter C, Gerner-Smidt P, Gilmour MW, et al. (2012) Integrating genome-based informatics to modernize global disease monitoring, information sharing, and response. Emerg Infect Dis 18: 1.
- World Health Organization (2018) Whole genome sequencing for foodborne disease surveillance: Landscape paper. World Health Organization, Geneva, Switzerland.
Citation: Schlundt J, Tay MYF (2020) The Revolutionizing Prospects of Next Generation DNA Sequencing of Microorganisms for Microbiological and AMR Identification and Characterization-the GMI Singapore Sequencing Statement. J Food Sci Nutr 6: 072.
Copyright: © 2020 Joergen Schlundt, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

