
Profiling the Secretome of Space Traveler Human Neural Stem Cells
*Corresponding Author(s):
Juan Carlos BiancottiDepartment Of Surgery, Division Of Pediatric Surgery, School Of Medicine, Johns Hopkins University, 1721 E. Madison St, Ross 733, Baltimore, MD 21205, United States
Tel:1+ 4432876656,
Email:jbianco5@jh.edu
Abstract
Graphical Abstract
Microgravity and the space environment have significant impact on the physiology of organisms. Understanding the responses elicited in the Central Nervous System (CNS) such as alteration in the visual function process and the regulation of biological pathways leading to changes of the CNS function, is critical to design contra measures aiming to mitigate the impact on astronaut’s health. Here, we investigated the secretome of human neural stem cells cultured aboard the International Space Station for 39.3 days and compared it to the secretome of sister cells cultured in parallel on Earth. Examination of their secretome uncovered activation of stress responses among which are oxidative stress, endoplasmic reticulum stress, and proteotoxic stress responses. These results provide additional evidence on the impact that microgravity environment has on neural tissue and in particular neural stem cells, and potential impairment of the brain function.
Keywords
Microgravity; Neural stem cells; Proteomics; Secretome; Spaceflight; Stress responses
Introduction
Despite the increase in the number of studies, the effects of the space environment on the central nervous system (CNS) are not yet fully understood. Changes in gravitational forces experienced during spaceflights or simulated on Earth are challenging situations for cells and whole organisms. It has been reported that astronauts who returned from missions on the International Space Station presented with health problems, including alterations of the CNS, particularly in cerebellar, sensorimotor, and vestibular brain areas [1,2]. Studies in cell cultures and organisms revealed that cell cultures are more sensitive to microgravity (µG) than cells as part of an organism [3]. This high sensibility demonstrated by cell cultures may even represent an advantage to understand the effects of µG on individual organs. Using these systems, the most commonly reported alterations are inhibition of differentiation and retention of stemness, proliferation, survival, and oxidative stress [4-11]. Our own studies and those from other groups have shown increase in cell proliferation and metabolic activity of human Neural Stem Cells (NSCs) in a spaceflight or simulated µG, due in part to increase in mitochondria function [12,13]. In addition, protein synthesis and secretion are influenced by exposure to µG [14,15].
In this study, we investigate the effects of the space environment on the proteome of human NSCs flown to the space and cultured for 39.3 days aboard the ISS. Our results indicate activation of different mechanisms supporting survival and homeostasis in response to the stress produced by spaceflight, and downregulation in the expression of proteins involved in visual function and CNS development.
Methods
Cells
A homogeneous population of NSCs was obtained from human-induced Pluripotent Stem Cells (hiPS). The original cells, known as “CS83iCTR-33nxx” (such as skin cells), were “reprogrammed” and provided to us by Cedars-Sinai Medical Center via a material transfer agreement. For space flight, NSCs were attached to mesh carriers (2mm x 3mm) and placed inside a Type IV cell culture chamber (Airbus-Yuri, Friedrichshafen) that allows for media change at the docking to the ISS and before undocking to flight back to the Earth. The culture chambers were installed in the Space Technology and Advanced Research System Experiment Facility-1 at 37°C. After splash-down, samples were transported in a controlled environment. Upon arrival to UCLA, NSCs were retrieved from the hardware, plated onto poly-d-lysine coated flasks in our proprietary stem cell chemically defined medium (STM) [16,17] and allowed to recover from space flight and splash-down.
Secretome collection
The culture medium that fed the cells during space flight was recovered from the hardware separately, medium from the cell chamber, and from each tank were placed in numbered tubes with addition of proteases inhibitor cocktail, and saved frozen at -80°C. This medium is commonly known as the conditioned medium. For the purpose of this manuscript, we named this conditioned medium “secretome.”
Space flight information
NSCs were sent to space onboard SpaceX-16 using the Automated Type IV hardware from Yuri, intended to collect the secretome of NSCs during the stay at the ISS, removing the in flight media. When arriving to the ISS, the culture medium from the first tank is pumped into the cell chamber and incubated at 37°C. Before undocking, media is replaced by tank 2and the units were stored at 4ºC (Figure 1).
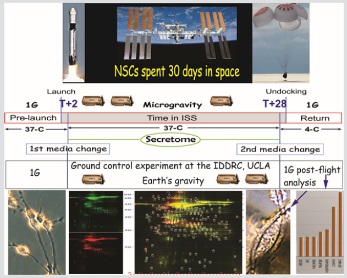
 Figure 1: Spaceflight automated experiment. As part of the Bioscience-4 Space Biology NASA experiment, flew to the ISS on board of SpaceX-16. The cells remained in microgravity for approximately 39.3 days, on board the International Space Station in an orbit of 354 miles above Earth. A) Shows the timeline for the spaceflight experiment. Following 2nd media change, the replaced medium containing the secretome was stored at 4°C until return to Earth. B) View of the Type IV chamber where cells were seeded and cultured. C) Bottom view of the Type IV unit showing the two tanks. Tank 1 medium is used for the 1st media change and Tank 2 medium was pumped into the cell chamber during the 2nd media change. D) View of the inner portion of facility STaARS-1EF, where units were installed once contained in their black external cover. Cells travelled and were incubated in this facility while in space. The STaARS-1 EF was designed by STaARS, inspired on this study having in mind the use of minimal astronaut time. Therefore, this facility is activated and controlled from STaARS’ headquarters in Houston, TX. A total of four Automated Type IV units were flown aboard SpaceX-16 on December 5, 2018. To view more details of the hardware please visit: https://www.yurigravity.com/our-service - right-hardware. E) View of NSCs growing at Cape Canaveral prior to seeding them onto the flying hardware.
Figure 1: Spaceflight automated experiment. As part of the Bioscience-4 Space Biology NASA experiment, flew to the ISS on board of SpaceX-16. The cells remained in microgravity for approximately 39.3 days, on board the International Space Station in an orbit of 354 miles above Earth. A) Shows the timeline for the spaceflight experiment. Following 2nd media change, the replaced medium containing the secretome was stored at 4°C until return to Earth. B) View of the Type IV chamber where cells were seeded and cultured. C) Bottom view of the Type IV unit showing the two tanks. Tank 1 medium is used for the 1st media change and Tank 2 medium was pumped into the cell chamber during the 2nd media change. D) View of the inner portion of facility STaARS-1EF, where units were installed once contained in their black external cover. Cells travelled and were incubated in this facility while in space. The STaARS-1 EF was designed by STaARS, inspired on this study having in mind the use of minimal astronaut time. Therefore, this facility is activated and controlled from STaARS’ headquarters in Houston, TX. A total of four Automated Type IV units were flown aboard SpaceX-16 on December 5, 2018. To view more details of the hardware please visit: https://www.yurigravity.com/our-service - right-hardware. E) View of NSCs growing at Cape Canaveral prior to seeding them onto the flying hardware.
2-D DIGE
- Preparation of samples
The cell culture medium was collected from the hardware, protease inhibitor cocktail added and then stored at -80°C. To begin the analysis, samples were thawed, vortexed for 20 sec, spun down for 30 min at 14,000 rpm at 4°C, and the supernatant was collected. For the two NSC samples, serum albumin and IgG were removed using Thermo Scientific Albumin/IgG Removal Kit. Next, the depleted serum samples were concentrated and exchanged into 2-D Lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, and 30 mM Tris-HCl, pH 8.5). Protein concentration was measured in all samples using Bio-Rad protein assay method.
The reaction was stopped by adding 1.0 µl of 10 mM Lysine to each sample, and incubating in the dark on ice for additional 15 min. The labeled samples were then mixed. The 2X 2-D Sample buffer (8 M urea, 4% CHAPS, 20 mg/ml DTT, 2% pharmalytes and trace amount of bromophenol blue), 100 ul destreak solution and Rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 20 mg/ml DTT, 1% pharmalytes and trace amount of bromophenol blue), were added to the labeling mix to make the total volume of 250 µl. Mixing well and spinning before loading the labeled samples into the strip holder.
- IEF and SDS-PAGE
After loading the labeled samples, IEF (pH3-10 Non-Linear) was run following the protocol provided by GE Healthcare. Upon finishing the IEF, the IPG strips were incubated in the freshly made equilibration buffer-1 (50 mM Tris-HCl, pH 8.8, containing 6 M urea, 30% glycerol, 2% SDS, trace amount of bromophenol blue and 10 mg/ml DTT), for 15 minutes with gentle shaking. Then the strips were rinsed in the freshly made equilibration buffer-2 (50 mM Tris-HCl, pH 8.8, containing 6 M urea, 30% glycerol, 2% SDS, trace amount of bromophenol blue and 45 mg/ml Iodoacetamide), for 10 minutes with gentle shaking. Next, the IPG strips were rinsed in the SDS-gel running buffer before transferring into 13.5% SDS-gels. The SDS-gels were run at 15°C until the dye front ran out of the gels.
- Image scan and data analysis
Gel images were scanned immediately following the SDS-PAGE using Typhoon TRIO (GE Healthcare). The scanned images were then analyzed by Image Quant software (version 6.0, GE Healthcare), followed by in-gel analysis using DeCyder software (version 5.0, GE Healthcare). The fold change of the protein expression levels was obtained from in-gel DeCyder analysis (Supplementary Figure 1).
Protein identification by mass spectrometry
- Spot picking and trypsin digestion
The spots of interest were picked up by Ettan Spot Picker (AmershamBioSciences) based on the in-gel analysis and spot picking design by DeCyder software. The gel spots were washed a few times then digested in-gel with modified porcine trypsin protease (Trypsin Gold, Promega). The digested tryptic peptides were desalted by Zip-tip C18 (Millipore). Peptides were eluted from the Zip-tip with 0.5 ul of matrix solution (α-cyano-4-hydroxycinnamic acid (5 mg/ml in 50% acetonitrile, 0.1% trifluoroacetic acid, 25 mM ammonium bicarbonate)) and spotted on the AB SCIEX MALDI plate (Opti-TOFTM 384 Well Insert).
- Mass Spectrometry
MALDI-TOF MS and TOF/TOF tandem MS/MS were performed on an AB SCIEX TOF/TOF™ 5800 System (AB SCIEX, Framingham, MA). MALDI-TOF mass spectra were acquired in reflectron positive ion mode, averaging 4000 laser shots per spectrum. TOF/TOF tandem MS fragmentation spectra were acquired for each sample, averaging 4000 laser shots per fragmentation spectrum on each of the 10 most abundant ions present in each sample (excluding trypsin autolytic peptides and other known background ions).
- Database search
Both the resulting peptide mass and the associated fragmentation spectra were submitted to the GPS Explorer workstation equipped with the MASCOT search engine (Matrix Science) to search the database of the National
Center for Biotechnology Information non-redundant (NCBInr) or Swiss-Prot-database.
Searches were performed without constraining protein molecular weight or isoelectric point, with variable carbamidomethylation of cysteine and oxidation of methionine residues. One missed cleavage was also allowed in the search parameters. Candidates with either protein score C.I.% or Ion C.I.% greater than 95 were considered significant.
The list of proteins with levels SPC-NSCs/GC-NSC ≥ 2 or ≤ -2 was loaded into the Reactome Pathway database. Reactome is a curated and peer-reviewed database of pathways and reactions in human biology [18] https://reactome.org/PathwayBrowser/#/ANALYSIS=MjAyMDA3MDcxNjE4NTNfMjIyMzc%3D). For the analysis, we have considered only those pathways with a probability score, corrected for false discovery rate by the Benjamini-Hochberg method, < 0.05.We use EnrichR to predict transcription factors regulating proteins with levels SPC-NSCs/GC-NSC ≥ 1.3.
Results
- Space-Dependent protein changes
Upon arrival to the laboratory after splash down, NSCs were found attached along the mesh fibers, and in some cases forming spheres adhered to the substrate (Figure 2). NSCs were retrieved from the hardware and plated onto poly-D-lysine coated flasks and allow to recover.
 Figure 2: Views of the mesh carrier culture system. A) To prevent cells from detaching from the surface of the hardware, during ascent and descent, cells were pre-seeded onto mesh carriers (2mm × 3mm). The arrow shows the mesh where cells were seeded. B) NSCs grew along the mesh fibers and in some cases formed spheres that remained adhered to the substrate. C) NSCs also grew across the spaces left by the mesh arranging themselves in a tissue-like manner. This was advantageous as it maximized the surface for cell growth. B and C: 200x magnification.
Figure 2: Views of the mesh carrier culture system. A) To prevent cells from detaching from the surface of the hardware, during ascent and descent, cells were pre-seeded onto mesh carriers (2mm × 3mm). The arrow shows the mesh where cells were seeded. B) NSCs grew along the mesh fibers and in some cases formed spheres that remained adhered to the substrate. C) NSCs also grew across the spaces left by the mesh arranging themselves in a tissue-like manner. This was advantageous as it maximized the surface for cell growth. B and C: 200x magnification.
A total of 119 differentially expressed proteins were identified by analysis of the secretome of NSCs, 86 upregulated in SPC-NSCs/GC-NSCs and 34 downregulated. From those, 37 upregulated proteins have a fold change ≥ 2 and 5 downregulated proteins a fold change ≤ -2 (Figure 3). Figure 3: Most significantly increased and decreased proteins in spaceflight NSC cultures. Proteins released by NSCs after 39.3 days in space onboard the ISS. Bars to the right depicts proteins found to be ≥ 2 in SPC-NSCs secretome compared to GC-NSCs. Bars on the left are proteins in lower amounts (≤ -2) in SPC-NSCs secretome compared to GC-NSCs. Significance p < 0.05.
Figure 3: Most significantly increased and decreased proteins in spaceflight NSC cultures. Proteins released by NSCs after 39.3 days in space onboard the ISS. Bars to the right depicts proteins found to be ≥ 2 in SPC-NSCs secretome compared to GC-NSCs. Bars on the left are proteins in lower amounts (≤ -2) in SPC-NSCs secretome compared to GC-NSCs. Significance p < 0.05.
- Space travel triggers stress responses in NSCs
The proteomic analysis of the secretome produced by SPC-NSCs and GC-NSCs revealed an overall increase in secreted proteins by SPC-NSCs involved in stress response and autophagy. For example, “Chaperone mediated autophagy”, “Detoxification of Reactive Oxygen Species (ROS)”, “ATF6 activation of chaperone genes”, and heat shock transcription factor 1 (HSF1) activation” are among the most significantly activated pathways in SPC-NSCs (Table 1). In line with the concept of cellular stress/damage, the levels of proteins such as peroxiredoxin-1 (PRDX1), thioredoxin-dependent peroxide reductase (PRDX3), and protein disulfide-isomerase (P4HB), involved in quenching of ROS, are 2.1 and 2.2 times higher in SPC-NSCs culture medium than in GC-NSCs control medium (Figure 3; Suppl. Table 1). We found changes in proteins implicated in activation of endoplasmic reticulum (ER) stress response, Secreted Protein Acidic and Cysteine Rich (SPARC), Calreticulin (CALR) and Endoplasmin (HSP90B1) are found to be 12.8, 5.2 and 3.3 times, respectively, more expressed in SPC-NSCs than GC-NSCs (Figure 3; Suppl. Table 1).The marked increased levels of SPARC in SPC-NSCs flown cells led us to examine these cells using time-lapse microscopy. We have found the occurrence of autophagy in NSC cultures exposed to space conditions (Study in progress). The levels of two members of the heat shock factor 1 (HSF1)pathway, 14-3-3ε (YWHAE) and HSP90B (HSP90AB1) proteins, were found to have increased by 2.1 and 7.3 folds in the secretome from SPC-NSCs (Figure 3; Suppl. Table 1).
|
Pathway name |
Entities |
Reactions |
||||
|
found |
ratio |
p-value |
FDR |
found |
ratio |
|
|
Chaperone Mediated Autophagy |
3/22 |
0.002 |
4.08e-05 |
0.01 |
15/19 |
0.001 |
|
ECM proteoglycans |
4/76 |
0.007 |
7.73e-05 |
0.01 |
4/23 |
0.002 |
|
Detoxification of Reactive Oxygen Species |
3/39 |
0.003 |
2.20e-04 |
0.014 |
3/34 |
0.003 |
|
Post-translational protein phosphorylation |
4/107 |
0.009 |
2.85e-04 |
0.014 |
1/1 |
7.63e-05 |
|
Interleukin 12 signaling |
3/46 |
0.004 |
3.55e-04 |
0.014 |
4/56 |
0.004 |
|
ATF6 (ATF6-alpha) activates chaperone genes |
2/10 |
8.75e-04 |
4.22e-04 |
0.014 |
2/5 |
3.81e-04 |
|
Regulation of IGF transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
4/124 |
0.011 |
4.95e-04 |
0.014 |
1/14 |
0.001 |
|
Binding and Uptake of Ligands by Scavenger Receptors |
4/129 |
0.011 |
5.74e-04 |
0.014 |
7/33 |
0.003 |
|
HSF1 activation |
2/12 |
0.001 |
6.05e-04 |
0.014 |
4/7 |
5.34e-04 |
|
ATF6 (ATF6-alpha) activates chaperones |
2/12 |
0.001 |
6.05e-04 |
0.014 |
2/10 |
7.63e-04 |
|
Interleukin 12 family signaling |
3/56 |
0.003 |
6.28e-04 |
0.014 |
5/114 |
0.009 |
|
Attenuation phase |
2/14 |
0.001 |
8.20e-04 |
0.016 |
4/5 |
3.81e-04 |
|
Scavenging by Class A Receptors |
2/19 |
0.002 |
0.001 |
0.028 |
2/10 |
7.63e-04 |
|
Extracellular matrix organization |
5/301 |
0.026 |
0.002 |
0.029 |
20/319 |
0.024 |
|
Neurodegenerative Diseases |
2/22 |
0.002 |
0.002 |
0.029 |
2/22 |
0.002 |
|
Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer’s disease models |
2/22 |
0.002 |
0.002 |
0.029 |
2/22 |
0.002 |
|
Signaling by Interleukins |
6/456 |
0.04 |
0.002 |
0.029 |
8/493 |
0.038 |
|
HSF1-dependent transactivation |
2/24 |
0.002 |
0.002 |
0.031 |
5/8 |
6.10e-04 |
|
Cellular response to heat stress |
3/95 |
0.008 |
0.003 |
0.033 |
17/29 |
0.002 |
|
TALDO1 deficiency: failed conversion of Fru (6), E4P to SH7P, GA3P |
1/1 |
8.75e-05 |
0.003 |
0.033 |
1/1 |
7.63e-05 |
|
Diseases of programmed cell death |
2/29 |
0.003 |
0.003 |
0.038 |
2/24 |
0.002 |
|
Interleukin-4 and Interleukin-13 signaling |
3/111 |
0.01 |
0.004 |
0.044 |
3/47 |
0.004 |
|
Late endosomal microautophagy |
2/34 |
0.003 |
0.005 |
0.047 |
3/3 |
2.29e-04 |
|
Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation |
2/37 |
0.003 |
0.005 |
0.048 |
2/36 |
0.003 |
Table 1: Most significantly upregulated pathways in SPC-NSCs. This table shows the 25 most relevant pathways sorted by p-value obtained from an input list of proteins found to be ≥ 2 times more abundant in SPC-NSCs compared with GC-NSCs. The data was generated using Reactome Pathway database.
In addition to the above-mentioned pathways, we identified activation of IL-12 signaling, which is involved in Th1 proinflammatory cell response. Three members of this pathway, MIF, TALDO and PDIA1 (P4HB) were significantly upregulated by 2.1 - 5.3 times in SPC-NPCs culture medium. L-lactate dehydrogenase (LDHA) is an enzyme found in nearly all living cells and its release to the medium is commonly used as a marker of cell damage/death. LDHA protein content in SPC-NSC medium is 10.7 times higher than in ground control cells and may be indication of cell damage and/or cell death.
Based on the secretome data, we could infer that some transcription factors upstream of these differentially expressed proteins contributed to the proteomics profile. EnrichR (ENCODE and ChEA consensus) identified MYC and NELFE as candidate transcription factors regulating proteins more expressed in SPC-NSCs than GC-NSCs (Table 2). Due to the shorter list of proteins reduced in SPC-NSCs compared to GC-NPCs, no significant transcription factors could be identified (based on adjusted P-value).
|
Term |
p-value |
Adjusted p-value |
Combined score |
|
NELFE |
6.61e-08 |
5.22e-06 |
296.0 |
|
MYC |
5.07e-05 |
0.002004237 |
69.6 |
|
ZMIZ1 |
0.00118812 |
0.031287085 |
29.0 |
|
CREB1 |
0.00164278 |
0.032444952 |
22.4 |
Table 2: Transcription factors related to the upregulated proteins. Transcription factors predicted to be upstream of an input list of proteins found to be ≥ 1.3 times more abundant in SPC-NSCs compared with GC-NSCs. Data obtained from EnrichR (ENCODE and ChEA consensus).
Overall, the analysis of the secretome of NSCs indicates that exposure to the space environment induces stress responses as a mean to cope with changes in gravitational forces.
- Space travel affects proteins involved in vision and neural development
A few proteins were found significantly reduced in SPC-NSC with respect to GC-NSC secretome. Among them, Calmodulin-2 (CALM2) and Transthyretin (TTR) come up as the two most significant proteins based on their involvement in numerous biological pathways. CALM2 was found reduced by 6.1 times in SPC-NSCs secretome, and the most significantly associated pathways include the metabolism of vitamins and cofactors related to visual phototransduction, CREB1 phosphorylation through activation of CaMKK and adenylate cyclase, and loss of phosphorylation of MECP2, which conduce to disorders of the nervous system development (Table 3 and Suppl. Table 1).
|
Pathway name |
Entities |
Reactions |
||||
|
found |
ratio |
p-value |
FDR |
found |
ratio |
|
|
Visual phototransduction |
2/100 |
0.009 |
0.001 |
0.029 |
6/92 |
0.007 |
|
Neutrophil degranulation |
3/480 |
0.041 |
0.001 |
0.029 |
3/10 |
7.33e-04 |
|
Innate Immune System |
4/1,191 |
0.103 |
0.001 |
0.029 |
7/710 |
0.052 |
|
Cam-PDE 1 activation |
1/4 |
3.45e-04 |
0.002 |
0.029 |
1/2 |
1.47e-04 |
|
Loss of phosphorylation of MECP2 at T308 |
1/5 |
4.31e-04 |
0.003 |
0.029 |
1/1 |
7.33e-05 |
|
Activation of RAC1 downstream of NMDARs |
1/7 |
6.04e-04 |
0.004 |
0.029 |
5/6 |
4.40e-04 |
|
Metabolism of vitamins and cofactors |
2/192 |
0.017 |
0.004 |
0.029 |
3/206 |
0.015 |
|
CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascade |
1/8 |
6.90e-04 |
0.004 |
0.029 |
8/10 |
7.33e-04 |
|
Calcineurin activates NFAT |
1/9 |
7.77e-04 |
0.005 |
0.029 |
2/3 |
2.20e-04 |
|
Activation of Ca-permeable Kainate Receptor |
1/10 |
8.63e-04 |
0.005 |
0.029 |
2/2 |
1.47e-04 |
|
CaMK IV-mediated phosphorylation of CREB |
1/10 |
8.63e-04 |
0.005 |
0.029 |
10/13 |
9.53e-04 |
|
Ionotropic activity of kainate receptors |
1/10 |
8.63e-04 |
0.005 |
0.029 |
2/4 |
2.93e-04 |
|
Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
1/10 |
8.63e-04 |
0.005 |
0.029 |
2/16 |
0.001 |
|
CLEC7A (Dectin-1) induces NFAT activation |
1/11 |
9.49e-04 |
0.006 |
0.029 |
4/6 |
4.40e-04 |
|
eNOS activation |
1/11 |
9.49e-04 |
0.006 |
0.029 |
9/20 |
0.001 |
|
CREB1 phosphorylation through the activation of Adenylate Cyclase |
1/12 |
0.001 |
0.006 |
0.029 |
2/6 |
4.40e-04 |
|
Reduction of cytosolic Ca++ levels |
1/12 |
0.001 |
0.006 |
0.029 |
1/3 |
2.20e-04 |
|
Pervasive developmental disorders |
1/13 |
0.001 |
0.007 |
0.029 |
1/5 |
3.66e-04 |
|
Loss of function of MECP2 in Rett syndrome |
1/13 |
0.001 |
0.007 |
0.029 |
1/5 |
3.66e-04 |
|
Disorders of Nervous System Development |
1/13 |
0.001 |
0.007 |
0.029 |
1/5 |
3.66e-04 |
|
Disorders of Developmental Biology |
1/13 |
0.001 |
0.007 |
0.029 |
1/5 |
3.66e-04 |
|
Retinoid cycle diseases events |
1/13 |
0.001 |
0.007 |
0.029 |
1/16 |
0.001 |
|
Diseases of the neuronal system |
1/13 |
0.001 |
0.007 |
0.029 |
1/17 |
0.001 |
|
Disease associated with visual transduction |
1/13 |
0.001 |
0.007 |
0.029 |
1/17 |
0.001 |
Table 3: Most significantly downregulated pathways in SPC-NSCs. List of the 25 most relevant pathways sorted by p-value from the list of significantly decreased proteins in SPC-NSCs in comparison to GC-NSCs. The data was generated using Reactome Pathway database.
In the SPC-NSCs secretome TTR protein was found reduced 4.6 times with respect to GC-NSCs (Figure 3 and Suppl. Table 1). The most significantly involved pathways were related to retinol cycle diseases including visual phototransduction, and disorders of developmental biology, in particular of the neural system (Table 3). Two additional downregulated proteins in the SPC-NSCs secretome were WNT2B and GPI (glucose-6-phosphate isomerase), by 2.2 and 2.1 folds, respectively (Figure 3 and Suppl. Table 1). Overall, downregulation of these secreted proteins in the SPC-NSCs culture medium suggest that spaceflight may impair the visual function process and the regulation of different biological pathways, leading to alterations of the nervous system.
Discussion
In this study, we investigated the effect of the space environment on hiPS-derived NSCs cultures maintained in the ISS for 39.3 days. Utilizing a proteomics approach, we analyzed the secretome of NSCs in space and on Earth to uncover insights into biological mechanisms of adaptation to space microgravity. Our analyses reveal activation of different stress response pathways in spaceflight cell cultures suggesting that these are triggered adaptation mechanisms in response to the environmental challenge aiming to regain homeostasis. Exposure to a µG environment, by either spaceflight or simulated on-ground-based models, leads to oxidative damage to different tissues in humans and animals, due to generation of excessive reactive oxygen/reactive nitrogen species (ROS/RNS) [8,9,19], which may even occur after returning to Earth. For example, urinary excretion of the oxidative damage markers, 8-iso-prostaglandin F2α and 8-hydroxydeoxyguanosine were measured in-flight (88 to 186 days in orbit) and post-flight (up to 14 days) in the Mir mission crew [20]; alterations in mitochondrial metabolites and mtDNA were observed in both mouse models and humans in a multi-omics study [9]; transcriptional activation of genes involved in ROS metabolism and cellular response to oxidative stress were detected in human myelomonocytic U937 cells using a parabolic flight and suborbital ballistic rocket experiments [7]. In line with previous reports, we observed a significant increase of proteins from the “Detoxification of ROS” metabolic pathway in the secretome of SPC-NSCs, supporting the notion that NSCs also experience oxidative stress when exposed to space conditions of µG.
The analysis of the secretome also revealed activation of additional biologic pathways related to stress and immune response, such as “HSF1 activation,” “cellular response to heat stress,” and “IL12 activation.” In particular, “ATF6-α activates chaperone genes” pathway is directly related to ER stress, as ATF6-αtogether with the transcription factor NF-Y bind to ER Stress Response Elements activating transcription of ER stress-responsive genes [21]. Our secretome analysis supports this line of observations, as the levels of proteins involved in ER stress and members of the “ATF6 activation of chaperone genes” and “chaperone-mediated autophagy” pathways are significantly increased in the SPC-NSCs culture medium. CALR and HSP90B1 are resident chaperones of the ER, acting as downstream intermediates from the activation of ATF6-alpha. They bind to misfolded proteins and prevent them from being exported to the Golgi apparatus [22]. These observations also indicate that ER stress and autophagy occur not only in simulated µG but also in the space environment.
Autophagy is an evolutionary conserved lysosome-mediated process enabling degradation and recycling of dysfunctional proteins or whole organelles. It is an essential degradative pathway for the homeostasis of tissues, especially the CNS, and a means cells use to cope with stress/damage [23,24]. It may either result in cell survival or lead to cell death, depending on the type and magnitude of the stress/damage [25]. SPARC is involved in numerous processes that induce endoplasmic reticulum stress, which eventually leads to autophagy [26]. Recent studies have provided evidence suggesting that simulated µG using random positioning machines (RPMs) activate autophagy in RAW264.7 pre-osteoclast cells and seminoma cells [27,28]. It has been recently shown that autophagy protects against µG-induced ER stress in HUVEC cells [29]. Interestingly, ground control cells also displayed this behavior implying that having been unattended for 45 days prompted this stress response that continued after cells were recovered from the flying hardware and fed with fresh medium before time-lapse acquisition. A subsequent space flight would be necessary to obtain more NSCs to study changes of mechanisms and pathways exerted on these cells by space flight.
In addition to the above mentioned stress pathways, a more general stress response pathway, the activation of heat shock factor 1 (HSF1), implicates the activation of gene expression in response to a variety of stresses, including heat shock, oxidative stress, as well as inflammation and infection [30-33]. Altogether, the activation of these pathways strengthens the notion of a generalized challenge imposed by changes in gravity conditions to NSCs. It is now clear that mammalian cells sense gravitational alterations as a stressful event and turn physical cues into biochemical signals, which reprogram their activities [34].
The few proteins that were reduced in SPC-NSCs culture medium have significant implications in cell physiology. For example, CALM2, a calcium-binding protein, plays a role in signaling pathways, cell cycle progression and proliferation [35,36]. Through its calcium-binding property it regulates a large number of enzymes (kinases, phosphatases), ion channel, and aquaporins, among others. TTR is a carrier protein involved in the transport of retinol (vitamin A) in the plasma by association with retinol-binding protein, in nerve regeneration, autophagy, and proteolysis [37-39]. In the mammalian brain, TTR is produced and secreted by choroid plexus epithelial cells where it binds to thyroid hormone (TH) and is distributed via the CSF. The complex TTR-TH reaches the subventricular zone (SVZ) and directs the specification of NSCs towards either a neuronal or glial phenotype. It has been shown that lack of TTR expression results in a decreased neuronal production with a concomitant increase in oligodendrocyte progenitor fate choice. There are also gender specific differences with respect to which region of the SVZ is being studied [40]. This is important as a reduction in neurons may lead to cognitive decline. WNT2B plays a critical role in development, including regulation of cell growth and differentiation, whereas GPI as an extracellular protein, functions as a neurotrophic factor in addition to its role in the cytoplasmic glycolytic pathway [41]. The Wnt signaling pathway is associated with proper neuronal connectivity and synapses. Wnts are expressed in the brain since early in development and persist in adulthood where they remodel pre- and post-synaptic regions and therefore synaptic connectivity and function [42]. Therefore, a decrease in Wnts may lead to a poor renewal of hippocampal neurons. Altogether, this evidence suggests that lower levels of these proteins may compromise CNS physiology and function, providing new evidence on possible mechanisms for brain and cognitive deterioration, particularly in long spaceflight missions.
Ionizing Radiation (IR) and μG exposure act simultaneously in space, producing interactions between adaptive pathways and defense responses. Though activation of stress and inflammatory responses have also been reported in NSCs under simulated μG, and the exposition to IR is significantly reduced by the protective shield surrounding the ISS, the contributing effect of IR in spaceflights is still a matter of consideration especially in long-time missions. IR generates ROS/RNS giving rise to oxidative stress and damage [43]. NSCs are extremely sensitive to IR damage. It has been reported that autophagy in NSCs is a protective mechanism in preventing IR-induced apoptosis. Decreased autophagy by knocking-down Atg7 significantly increased the incidence of apoptosis in irradiated NSCs [44]. It has also been reported that in R. rubrum S1H, a low dose of ionizing radiation at a level where it does not reduce cell viability, induces significant transcriptome changes observed in just a few significant differentially expressed proteins [45]. Future studies need to address the effects of low IR and microgravity on the proteomic profile of NSCs.
Finally, the NSCs used in this study like all organisms living on Earth, have been born and through the years have evolved in 1G. Both, acceleration and mechanical load disappear in weightlessness. Therefore, cells sense for the first time “free-floating” a condition unknown to the cell and its components leading to indiscriminate or undefined stress response that in turn generates unpredictable and perhaps even instable results [46]. At the present time, we interpret these changes as “stress responses” because nobody has defined how to start identifying “non-stress” from cellular responses specific to weightlessness. These differences may be the key to the modulation of neurodegeneration both on Earth and in the cosmos. These observations suggest that IR maybe a contributing factor to the effects observed on spaceflight NSCs once they leave the protection of the atmosphere.
Conclusion
The outer space is a challenging environment for unicellular and multicellular organisms that require adaptation to survive in those exigent conditions. NSC cultures onboard the ISS experience unrecognized lack of gravity that made them elicit a variety of stress responses, among which oxidative stress and ER stress appear as the most significant activated pathways. Autophagy, as a survival mechanism, appears to be activated under space µGas it does at simulated µG on Earth, as it was revealed by the marked increase in the secretion of SPARC in SPC-NSCs and time-lapse microscopy performed in our laboratory (manuscript in preparation). Other signs denoting alteration of the cell’s homeostasis are given by reduction in the secretion of proteins important for growth and differentiation of neural cells, the regulation of enzymatic signaling and nerve regeneration, suggesting compromise of CNS physiology and function. In addition, the contribution of IR in space in eliciting stress responses on organisms, although significantly reduced by the water shield in the ISS, cannot be ignored and needs to be considered in the interpretation of the results.
Taken together, the presented evidence implies that exposure of NSCs to the space environment affects different biologic functions in neural cells, inducing adaptive responses, and that this model system helps understanding the behavior of neural cells in space.
Author’s Contribution
JCB prepared the samples for the study, contributed to the global proteomics analysis, prepared the manuscript; LV, JZ contributed to the proteomic analyses; AE-J designed the entire study, chose the appropriate hardware, performed the work at Kennedy Space Center, recovered cells from the space hardware upon their return to UCLA and contributed to the discussion of the manuscript.
Funding
We thank NASA Space Biology for Grant NNX15AB43G. The IDDRC Cell Culture Core is supported by NIH/NICHD grant number U54HD087101-05.
Acknowledgements
We thank The Space Biology Team at NASA AMES, Dr. Amy Gresser, Elizabeth Pane, and Medaya Torres for their help with the implementation of the study. The NASA Space Biology Project team at Ames Research Center Dr. D. Tomko, K. Sato, E. Taylor, ARC Space Biology Project Manager and the support personnel at the Space Station Processing Facility at Kennedy Space Center. Thanks to Karin Perkins, Diana Ly, and Space Biology support staff. We also thank Dr. Carlos Cepeda for fruitful discussions. Special thanks to Maria Birlem and Chriss Bruderrek from Yuri, and STaARS team members Tom Kyler, Craig Walton and BreAnne MacKenzie for supporting the flight implementation. Thanks to Uli Kuebler from Airbus Defense and Space, who introduced me to the flying hardware used for this study. We also thank Elida Escalante, for preparation of the mesh micro-carriers. Dorwin Birt for help with computer-related matters. We thank Dr. Stephen Cederbaum and Dr. Harley Kornblum for signature of the student’s contract.
Data Availability Statement
Additional data will be available upon request
Conflict of Interest
The authors declare no conflict of interest.
References
- Ombergen AV, Demertzi A, Tomilovskaya E, Jeurissen B, Sijbers J, et al. (2017) The effect of spaceflight and microgravity on the human brain. J Neurol264: 18-22.
- Strollo F, Gentile S, Strollo G, Mambro A, Vernikos J (2018) Recent Progress in Space Physiology and Aging. Front Physiol9: 1551.
- Grimm D, Pietsch J, Wehland M, Richter P, Strauch SM, et al. (2014) The impact of microgravity-based proteomics research. Expert Rev Proteomics11: 465-76.
- Zhang Y, Wang H, Lai C, Wang L, Deng Y(2013) Comparative proteomic analysis of human SH-SY5Y neuroblastoma cells under simulated microgravity. Astrobiology 13: 143-150.
- Blaber EA, Finkelstein H, Dvorochkin N, Sato KY, Yousuf R, et al. (2015) Microgravity Reduces the Differentiation and Regenerative Potential of Embryonic Stem Cells. Stem Cells Dev 24: 2605-2621.
- Yang X, Wang Y, Li Q, Zhong Y, Chen L, et al. (2018) The Main Molecular Mechanisms Underlying Methamphetamine- Induced Neurotoxicity and Implications for Pharmacological Treatment. Front Mol Neurosci11:186.
- Tauber S, Christoffel S, Thiel CS, Ullrich O (2018) Transcriptional Homeostasis of Oxidative Stress-Related Pathways in Altered Gravity. Int J Mol Sci 19: 2814.
- Luxton JJ, McKenna MJ, Lewis A, Taylor LE, George KA, et al. (2020) Telomere Length Dynamics and DNA Damage Responses Associated with Long-Duration Spaceflight. Cell Rep 33:108457.
- Silveira WA, Fazelinia H, Rosenthal SB, Laiakis EC, Kim MS, et al. (2020) Comprehensive Multi-omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 183: 1185-1201.
- Camberos V, Baio J, Mandujano A, Martinez AF, Bailey L, et al. (2021) The Impact of Spaceflight and Microgravity on the Human Islet-1+ Cardiovascular Progenitor Cell Transcriptome. Int J Mol Sci 22: 3577.
- Han Y, Zeger L, Tripathi R, Egli M, Ille F, et al. (2021) Molecular genetic analysis of neural stem cells after space flight and simulated microgravity on earth. Biotechnol Bioeng118: 3832-3846.
- Cepeda C, Vergnes L, Carpo N, Schibler MJ, Bentolila LA, et al. (2019) Human Neural Stem Cells Flown into Space Proliferate and Generate Young Neurons. Appl Sci (Basel) 9: 4042.
- Chiang MC, Lin H, Cheng YC, Yen CH, Huang RN, et al. (2012) beta-adrenoceptor pathway enhances mitochondrial function in human neural stem cells via rotary cell culture system. J Neurosci Methods 207: 130-136.
- Krüger M, Pietsch J, Bauer J, Kopp S, Carvalho DTO, et al. (2019) Growth of Endothelial Cells in Space and in Simulated Microgravity - a Comparison on the Secretory Level. Cell Physiol Biochem 52: 1039-1060.
- Pietsch J, Kussian R, Sickmann A, Bauer J, Weber G, et al. (2010) Application of free-flow IEF to identify protein candidates changing under microgravity conditions. Proteomics 10: 904-913.
- Jeffrey AE, Catania SGB, Zhao PM, Cole R, Edmond J, et al. (2002) Selective specification of CNS stem cells into oligodendroglial or neuronal cell lineage: cell culture and transplant studies. J Neurosci Res 69: 810-825.
- Jeffrey AE, Wakeman DR, Kim SU, Snyder EY, Vellis J, et al. (2009) Culture system for rodent and human oligodendrocyte specification, lineage progression, and maturation. Curr Protoc Stem Cell Biol.
- Jassal B, Matthews L, Viteri G, Gong C, Lorente P, et al. (2018) The Reactome Pathway Knowledgebase. Nucleic Acids Res 46: 649-655.
- Tahimic CGT, Globus RK (2017) Redox Signaling and Its Impact on Skeletal and Vascular Responses to Spaceflight. Int J Mol Sci 18: 2153.
- Stein TP (2000) The relationship between dietary intake, exercise, energy balance and the space craft environment. Pflugers Arch 441: 21-31.
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, et al. (2000) ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol 20: 6755-6767.
- Hillary RF, FitzGerald U (2018) A lifetime of stress: ATF6 in development and homeostasis. J Biomed Sci 25: 48.
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell l6: 463-477.
- Scrivo A, Bourdenx M, Pampliega O, Cuervo AM (2018) Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol 17: 802-815.
- Ondrej M, Cechakova L, Durisova K, Pejchal J, Tichy A (2016) To live or let die: Unclear task of autophagy in the radiosensitization battle. Radiother Oncol 119: 265-275.
- Sailaja GS, Bhoopathi P, Gorantla B, Chetty C, Gogineni VR, et al. (2013) The secreted protein acidic and rich in cysteine (SPARC) induces endoplasmic reticulum stress leading to autophagy-mediated apoptosis in neuroblastoma. Int J Onco l42: 188-196.
- Morabito C, Guarnieri S, Catizone A, Schiraldi C, Ricci G, et al. (2017) Transient increases in intracellular calcium and reactive oxygen species levels in TCam-2 cells exposed to microgravity. Sci Rep 7: 15648.
- Sambandam Y, Townsend MT, Pierce JJ, Lipman CM, Haque A, et al. (2014) Microgravity control of autophagy modulates osteoclastogenesis. Bone 61: 125-131.
- Li CF, Sun JX, Gao Y, Shi F, Pan YK, et al. (2018) Clinorotation-induced autophagy via HDM2-p53-mTOR pathway enhances cell migration in vascular endothelial cells. Cell Death Dis 9: 147.
- Shamovsky I, Nudler E (2008) New insights into the mechanism of heat shock response activation. Cell Mol Life Sci 5: 855-861.
- Akerfelt M, Morimoto RI, Sistonen L(2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11: 545-555.
- Björk JK, Sistonen L (2010) Regulation of the members of the mammalian heat shock factor family. FEBS J 277: 4126-4139.
- Anckar J, Sistonen L(2011) Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem 80:1089-1115.
- Najrana T, Esteban JS (2016) Mechanotransduction as an Adaptation to Gravity. Front Pediatr 4: 140.
- Kahl CR, Means AR (2003) Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr Rev 24: 719-736.
- Berchtold MW, Villalobo A (2014) The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim Biophys Acta 1843: 398-435.
- Goodman DS (1984) Vitamin A and retinoids in health and disease. N Engl J Med310: 1023-1031.
- Fleming CE, Saraiva MJ, Sousa MM (2007)Transthyretin enhances nerve regeneration. J Neurochem 103: 831-839.
- Teixeira CA, Almeida MR, Saraiva MJ (2016) Impairment of autophagy by TTR V30M aggregates: in vivo reversal by TUDCA and curcumin. Clin Sci (Lond) 130: 1665-1675.
- Vancamp P, Gothié JD, Luongo C, Sébillot A, Blay KL, et al. (2019) Gender-specific effects of transthyretin on neural stem cell fate in the subventricular zone of the adult mouse. Sci Rep 9: 19689.
- Guo Y, Wu J, Wang M, Wang X, Jian Y, et al. (2022) The metabolite saccharopine impairs neuronal development by inhibiting the neurotrophic function of glucose-6-phosphate isomerase. J Neurosci 42: 2631-2646.
- Oliva CA, Vargas JY, Inestrosa NC (2013) Wnt signaling: role in LTP, neural networks and memory. Ageing Res Rev 12: 786-800.
- Beheshti A, McDonald JT, Hada M, Takahashi A, Mason CE, et al. (2021) Genomic Changes Driven by Radiation-Induced DNA Damage and Microgravity in Human Cells. Int J Mol Sci 22: 10507.
- Shi W, Liu W, Ma J, Lu J, Yang X, et al. (2020) The role of Atg7-mediated autophagy in ionizing radiation-induced neural stem cell damage. Gene 738: 144485.
- Mastroleo F, Houdt RV, Leroy B, Benotmane MA, Janssen A, et al. (2009) Experimental design and environmental parameters affect Rhodospirillumrubrum S1H response to space flight. ISME J 3: 1402-1419.
- Grimm D, Wise P, Lebert M, Richter P, Baatout S (2011) How and why does the proteome respond to microgravity? Expert Rev Proteomics 8: 13-27.
Supplementary Data
|
NSC0G>1G |
Accession No. |
Gene |
Top Ranked Protein Name [Species] |
|
12.8 |
SPRC_HUMAN |
SPARC |
SPARC OS=Homo sapiens OX=9606 GN=SPARC PE=1 SV=1 |
|
10.7 |
LDHA_HUMAN |
LDHA |
L-lactate dehydrogenase A chain OS=Homo sapiens OX=9606 GN=LDHA PE=1 SV=2 |
|
7.3 |
HS90B_HUMAN |
HSP90AB1 |
Heat shock protein HSP 90-beta OS=Homo sapiens OX=9606 GN=HSP90AB1 PE=1 SV=4 |
|
7.0 |
NUCB1_HUMAN |
NUCB1 |
Nucleobindin-1 OS=Homo sapiens OX=9606 GN=NUCB1 PE=1SV=4 |
|
6.2 |
NCAM1_HUMAN |
NCAM1 |
Neural cell adhesion molecule 1 OS=Homo sapiens OX=9606 GN=NCAM1 PE=1 SV=3 |
|
6.2 |
VTDB_HUMAN |
GC |
Vitamin D-binding protein OS=Homo sapiens OX=9606 GN=GC PE=1 SV=1 |
|
6.2 |
GDIA_HUMAN |
GDI1 |
Rab GDP dissociation inhibitor alpha OS=Homo sapiens OX=9606 GN=GDI1 PE=1 SV=2 |
|
5.9 |
C1QBP_RAT |
|
Complement component 1 Q subcomponent-binding protein, mitochondrial OS=Rattus norvegicus OX=10116 |
|
5.3 |
MIF_HUMAN |
MIF |
Macrophage migration inhibitory factor OS=Homo sapiens OX=9606 GN=MIF PE=1 SV=4 |
|
5.2
|
CALR_HUMAN |
CALR |
Calreticulin OS=Homo sapiens OX=9606 GN=CALR PE=1 SV=1 |
|
4.8 |
NHA1_RHORH |
|
High-molecular weight cobalt-containing nitrile hydratase subunit alpha OS=Rhodococcus rhodochrous |
|
4.7 |
FXRD1_HUMAN |
FOXRED1 |
FAD-dependent oxidoreductase domain-containing protein1 OS=Homo sapiens OX=9606 GN=FOXRED1 PE=1SV |
|
4.7 |
PGCB_HUMAN |
BCAN |
Brevican core protein OS=Homo sapiens OX=9606 GN=BCAN PE=1 SV=2 |
|
4.2 |
LAMA2_HUMAN |
LAMA2 |
Laminin subunit alpha-2 OS=Homo sapiens OX=9606 GN=LAMA2 PE=1 SV=4 |
|
4.2 |
GLOD4_HUMAN |
GLOD4 |
Glyoxalase domain-containing protein 4 OS=Homo sapiens OX=9606 GN=GLOD4 PE=1 SV=1 |
|
4.2 |
K2C1_HUMAN |
KRT1 |
Keratin, type II cytoskeletal 1 OS=Homosapiens OX=9606 GN=KRT1 PE=1 SV=6 |
|
3.7 |
DZAN1_HUMAN |
DZANK1 |
Double zinc ribbon and ankyrin repeat-containing protein 1 OS=Homo sapiens OX=9606 GN=DZANK 1PE=1S
|
|
3.7 |
HSP7C_HUMAN |
HSPA8 |
Heat shock cognate 71 kDa protein OS=Homo sapiens OX=9606 GN=HSPA8 PE=1 SV=1 |
|
3.3 |
ENPL_HUMAN |
HSP90B1 |
Endoplasmin OS=Homo sapiens OX=9606 GN=HSP90B1 PE=1 SV=1 |
|
3.2 |
K1C10_HUMAN |
KRT10 |
Keratin, type I cytoskeletal 10 OS=Hom osapiens OX=9606 GN=KRT10 PE=1 SV=6 |
|
3.2 |
Z324A_HUMAN |
ZNF324 |
Zinc finger protein 324A OS=Homo sapiens OX=9606 GN=ZNF324 PE=2 SV=1 |
|
2.7 |
TALDO_HUMAN |
TALDO1 |
Transaldolase OS=Homo sapiens OX=9606 GN=TALDO1 PE=1 SV=2 |
|
2.5 |
AATM_HUMAN |
GOT2 |
Aspartate aminotransferase, mitochondrial OS=Homo sapiens OX=9606 GN=GOT2 PE=1 SV=3 |
|
2.5 |
UCHL1_HUMAN |
UCHL1 |
Ubiquitin carboxyl-termina hydrolase isozyme L1 OS=Homo sapiens OX=9606 GN=UCHL1PE=1 SV=2 |
|
2.4 |
LIPP_HUMAN |
PNLIP |
Pancreatic triacylglycerol lipase OS=Homo sapiens OX=9606 GN=PNLIP PE=1 SV=1 |
|
2.4 |
PP4R4_HUMAN |
PPP4R4 |
Serine/threonine-protein phosphatase 4 regulatory subunit 4 OS=Homo sapiens OX=9606 GN=PPP4R4 PE=1 |
|
2.2 |
PRDX3_HUMAN |
PRDX3 |
Thioredoxin-dependent peroxide reductase ,mitochondrial OS=Homo sapiens OX=9606 GN=PRDX3 PE=1 SV=3 |
|
2.1 |
MYOM1_HUMAN |
MYOM1 |
Myomesin-1 OS=Homo sapiens OX=9606 GN=MYOM1 PE=1 SV=2 |
|
2.1 |
K1C9_HUMAN |
KRT |
Keratin, type I cytoskeletal 9 OS=Homo sapiens OX=9606 GN=KRT9 PE=1 SV=3 |
|
2.1 |
VIME_HUMAN |
VIM |
Vimentin OS=Homo sapiens OX=9606 GN=VIM PE=1 SV=4 |
|
2.1 |
1433E_HUMAN |
YWHAE |
14-3-3 protein epsilon OS=Homo sapiens OX=9606 GN=YWHAE PE=1 SV=1 |
|
2.1 |
PRDX1_HUMAN |
PRDX1 |
Peroxiredoxin-1 OS=Homo sapiens OX=9606 GN=PRDX1 PE=1 SV=1 |
|
2.1 |
PDIA1_HUMAN |
P4HB |
Protein disulfide-isomerase OS=Homo sapiens OX=9606 GN=P4HB PE=1 SV=3 |
|
2.0 |
SETLP_HUMAN |
SETSIP |
Protein SETSIP OS=Homo sapiens OX=9606 GN=SETSIP PE=1 SV=1 |
|
NSC0G<1G |
Accession No. |
Gene |
Top Ranked Protein Name [Species] |
|
-6.2 |
K2C1_HUMAN |
KRT1 |
Keratin, type II cytoskeletal 1OS=Homo sapiens OX=9606 GN=KRT1 PE=1SV=6 |
|
-6.1 |
CALM2_HUMAN |
CALM2 |
Calmodulin-2 OS=Homo sapiens OX=9606 GN=CALM2 PE=1 SV=1 |
|
-4.6 |
TTHY_BOVIN |
TTR |
Transthyretin OS=Bos Taurus OX=9913 GN=TTR PE=1 SV=1 |
|
-2.2 |
WNT2B_HUMAN |
WNT2B |
Protein Wnt-2b OS=Homo sapiens OX=9606 GN=WNT2B PE=1 SV=2 |
|
-2.1 |
G6PI_HUMAN |
GPI |
Glucose-6-phosphate isomerase OS=Homo sapiens OX=9606 GN=GPI PE=1 SV=4 |
Supplementary Table 1: Most significant upregulated and downregulated proteins in SPC-NSCs. Proteins identified in the secretome of NSCs after 39.3 days in space onboard the ISS. Top part of the table lists significantly (p < 0.05) proteins found in amounts ≥ 2 in SPC-NSCs secretome compared to GC-NSCs. Bottom part shows proteins decreased in SPC-NSCs vs. GC-NSCs by ≤ -2 (p < 0.05).
Supplementary Figure 1: 2-D gels from GC-NSC and SPC-NSC secretomes and the overlap between both images. Gel images were scanned immediately following the SDS-PAGE using Typhoon TRIO (GE Healthcare). The scanned images were then analyzed by Image Quant software (version 6.0, GE Healthcare), followed by in-gel analysis using DeCyder software (version 5.0, GE Healthcare).
Supplementary data 1: Details of the SpaceX CRS-16 mission to the ISS
SpaceX CRS-16
Description
SpaceX CRS-16, also known as SpX-16, was a Commercial Resupply Service mission to the International Space Station launched on 5 December 2018 aboard a Falcon 9 launch vehicle. The mission was contracted by NASA and is flown by SpaceX. This CRS mission is the first with the Falcon 9 Block 5. Wikipedia
Berthing date: 8 December 2018, 15:36 UTC
Berthing port: Harmony nadir
Dry mass: 4,200 kg (9,300 lb)
Landing date: 14 January 2019, 05:10 UTC
Mission duration: 39 days, 10 hours, 54 minutes
Rocket: Falcon 9 Block 5
Spacecraft: Dragon C112.2
Supplementary data 2: Radiation dosage onboard the ISS during CRS-16 flight
Ionizing radiation on the ISS comes from two sources, Galactic Cosmic Radiation (GCR) and trapped protons, with most of the trapped proton dose accumulated during passages through the South Atlantic Anomaly (SAA).
The instruments that took these data, JSC-SRAG uses information on the ISS orbit combined with mapping of the geomagnetic field to break out the dose into GCR and SAA components.

Citation: Biancotti JC, Carpo N, Zamudio J, Vergnes L, Espinosa-Jeffrey A (2022) Profiling the Secretome of Space Traveler Human Neural Stem Cells. J Stem Cell Res Dev Ther 8: 094.
Copyright: © 2022 Juan Carlos Biancotti, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

