
A Rare Case of Dys385 Monoallelism Along With Amel Y Deletion: A Case Report
*Corresponding Author(s):
Ajay Kumar RanaDivision Of Biology, Central Forensic Science Laboratory Hyderabad, Ministry Of Home Affairs, Government Of India, Amberpet, Ramanthapur, Hyderabad, Telangana, India
Email:ajay1rana@gmail.com
Abstract
Y-chromosomal short tandem repeats analysis is commonly used in forensic science to determine paternity, genealogy and number of males in sexual assault cases. Due to the absence of a homologous Y chromosomal pairing partner, many mutations go unrepaired in the offspring. Here, we report a rare genetic case of monoallelism along with Y chromosome microdeletion in a paternity case. The paternity was established through DNA fingerprinting technique and statistical analysis of amplified alleles. In this case, rare Y microdeletion and DYS385 monoallelism were observed through Y-STR analysis. This case should focus not only on the rare interpretation of Y-haplotype profiles but also on the evolutionary loss or mutation of DNA segments from the Y chromosome, which does not affect virility/fertility.
Keywords
Forensic Genetics; Microdeletion; Monoallelism; Mutation; Paternity; Y-STR
Introduction
Y-STR profiling is highly useful in tracing of male patrilineage and unscrambling of multiple males in cases of murders or sexual assault [1]. The presence of male and female DNA in crime scene samples is usually distinguished by amplification of 112 bp and 106 bp from sex chromosomes (XY) [2]; thus, the male (XY) gives rise to two amplifications or peaks, while females (XX) generate a single amplification/peak. The maleness of an individual is determined by the testis-determining factor (TDF,~24 kDa protein) coded by the SRY gene (sex-determining region of the Y chromosome). Therefore, Y-chromosomal inheritance from father to male child is used to characterize paternal lineages of questioned or unknown male in paternity and sexual assault cases [3]. The acrocentric Y chromosome of humans is consisting of two unequal arms, a short arm (p) and a long arm (q), which are bounded by Pseudoautosomal Regions (PARs) at the tips [4] (Figure 1). The tip of the short arm, called PAR1 is~2.5 Mb while tip of the long arm PAR2 is ~320 kb, both ends also contain telomeric DNA [5]. These PARs are homologous regions also present on the X chromosome, and during meiosis, PARs behave like an autosome and recombine later after autosomes (delayed double strand break and pairing) [6]. Y-chromosomal inheritance from father to male child is used to characterise paternal lineages of questioned or unknown male trace donors, in paternity and sexual assault cases [3]. The sex determining gene SRY which codes for TDF is present in the PAR1 region of Y chromosome which is also translocated to X chromosome but very rarely (1 in~30000 of males) which gives rise to intersex condition called de la Chappelle Syndrome (XX, male syndrome) [7,8]. More than half of the Y chromosome (~40 Mb) is tightly packed heterochromatin-block present in long arm (Figure 1) [9,10], and very few protein coding genes (~70) are present in rest of the euchromatin portion [4], and it has been hypothesized that during evolution most of the genes have been relocated to other chromosomes or lost due to its specialization [11,12].
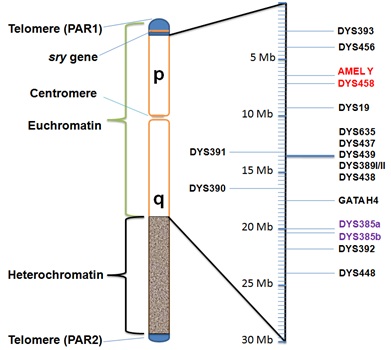 Figure 1: Location of AMEL Y and 17 Y-STRs in human Y chromosome.
Figure 1: Location of AMEL Y and 17 Y-STRs in human Y chromosome.
The amplified Y chromosomal AMEL Y and 17 Y-STRs have been positioned approximately along the Y chromosome along with other important markers like pseudoautosomal region (PAR) and sex-determining marker (SRY). The missing alleles in this case were located in consecutive positions along the short arm of the Y chromosome, with unusual detection of a monoallelic peak at the DYS385 locus.
The human Y chromosome pass from father to male offspring and is highly vulnerable to point mutations (~2x10-8 per base/generation) [13], STR mutations (~10-3 per locus/generation) [14,15], and microdeletions (~1:4000 men) [16] whereas errors in the DNA of other chromosomes are generally corrected by homologous chromosomal alignment and recombination from both parents during meiosis [17]. A remarkable recent finding demonstrates that as humans age and develop degenerative diseases, there is mosaic loss of Y-chromosome from blood cells [18, 19], which is further augmented by smoking habits, in fact up to four times greater than in non-smokers [20,21]. Here we report a rare case of monoallelic DYS385 STR along with microdeletion of short arm of Y chromosome. This case analysis is from a duo paternity case (a case of Yadav title surname belonging to peasant-pastoral communities, and consider themselves as the descendents of the King Yadu dynasty to which Lord Krishna belongs). The blood samples were collected by the medical officer on sterile gauze cloth pieces from the alleged father as well as the child as per direction from the court of law (The District and First Additional Session Judge cum Special Judge, Latehar, Jharkhand) and sent to the State Forensic Science Laboratory Jharkhand for DNA examination.
Materials and Methods
DNA extraction
The case exhibits (blood samples collected by the Authorized and Certified Medical Officer) of the alleged father and male child were provided to Director, State Forensic Science Laboratory Jharkhand on separate sterile gauze cloth pieces, and an area of approximately 1 cm2 was used for DNA isolation using the Trace DNA Purification Protocol (EZ1 DNA Investigator® Kit, Qiagen). The gauze pieces were placed in separate 2mL sterile sample tubes, and then 200µL of buffer G2 and 10 µL of proteinase K, were added. After brief vortexing (10s), the samples were mixed and placed in a water bath shaker set at 56oC for approximately 30 min. The sample tubes along with reagents cartridges, elution tubes and tip holders with tips were then loaded on the automated EZ1 worktable in their respective places (EZ1® Advanced XL, Qiagen). The purified DNA of the above samples were eluted shortly (~20 min) in TE buffer (40µL).
DNA quantification
The quantification of the above extracted DNA was carried out by real-time polymerase chain reaction (RT-PCR) using QuantifilerTM Duo DNA Quantification Kit (Applied Biosystems) [22]. Briefly, for every sample or standard DNA to be quantified, PCR Reaction Mix of 12.5 μL, Human Primer Mix of 10.5µL and sample/standard of 2µL were mixed in their respective tube of 96-Well Plate to obtain a final volume of 25 uL PCR reaction systems. The real time PCR machine (7500 Real Time PCR System, Applied Biosystems) calculated the quantity of DNA for each well as per standard DNA plot. Here, the DNA samples to be used as template were adjusted to 0.1ng/µL before setting up of PCR amplification.
Amplifications of STRs (Autosomal and Y) and genetic analysis
Fifteen autosomal STRs loci and one gender locus were co-amplified using AmpFLSTRTM Identifiler PlusTM PCR Amplification Kit (Table 1) [23]. This multiplexing was carried out in 0.2mL PCR tube using PCR Reaction Mix of 10 μL, Primer Set Mix of 5 μL and purified DNA of 10 μL (final volume of 25µL PCR reaction system). The amplification conditions were set to initial denaturation of 5 min at 95°C, followed by 28 cycles of 1 min denaturation at 94°C, 1 min annealing at 59°C and 1 min extension at 72°C, and lastly 10 min of final extension stage at 60°C. 1µL of these amplified PCR products or the AmpFlSTR® Identifiler Plus® allelic ladder were mixed to 8.7 μL of Hi-Di™ formamide and 0.3 μL of GeneScan™ 500 LIZ® Size Standard dye. This 10 μL electrophoresis system was subjected to 95oC for 5 min followed by snap chill (-20oC) for generating single stranded DNA forms. The samples were loaded on Genetic Analyzer (ABI-3130, Applied Biosystems) where capillary electrophoresis was performed in 36 cm 4-capillary array. Similarly multiplexing of the Y-STR alleles for the genomic DNA extracted was performed using AmpFLSTRTM Yfiler® PCR Amplification Kit [24]. The sizing of STR fragments separated in capillary to a single base resolution was carried out with GeneScanTM 500 LIZ® Size Standard and genetic analysis software GeneMapperTM ID v3.2. The observed resultant alleles size of the studied loci and their genotypes are represented in (Table 1) The whole experiment was repeated along with appropriate positive control (pooled human male genomic DNA as Standard DNA) and negative control (HiDiTM formamide) which were used during PCR amplification and are not shown here.
|
S. No. |
Autosomal-STR locus |
Alleged Father |
Child |
Paternity Index# |
Y-STR locus |
Alleged Father |
Child |
|
1. |
D8S1179 |
12, 14 |
12, 15 |
2.6315 |
DYS456 |
Allele not designated$ |
Allele not designated$ |
|
2. |
D21S11 |
28, 33.2 |
28, 29 |
1.8181 |
DYS389I |
12 |
12 |
|
3. |
D7S820 |
9, 12 |
9, 10 |
3.0303 |
DYS390 |
23 |
23 |
|
4. |
CSF1PO |
10, 11 |
10, 11 |
2.1897 |
DYS389II |
28 |
28 |
|
5. |
D3S1358 |
15, 16 |
15, 16 |
1.6597 |
DYS458 |
.... |
.... |
|
6. |
THO1 |
9, 9.3 |
6, 9 |
0.6944 |
DYS19 |
15 |
15 |
|
7. |
D13S317 |
8, 11 |
8, 12 |
0.9901 |
DYS385 |
13 |
13 |
|
8. |
D16S539 |
10, 13 |
9, 10 |
2.6315 |
DYS393 |
12 |
12 |
|
9. |
D2S1338 |
18, 19 |
18, 19 |
2.9286 |
DYS391 |
11 |
11 |
|
10. |
D19S433 |
13, 13 |
13, 13 |
3.1746 |
DYS439 |
11 |
11 |
|
11. |
vWA |
17, 18 |
15, 17 |
0.9091 |
DYS635 |
21 |
21 |
|
12. |
TPOX |
8, 9 |
9, 10 |
1.3513 |
DYS392 |
11 |
11 |
|
13. |
D18S51 |
12, 16 |
14, 16 |
1.7543 |
GATAH4 |
11 |
11 |
|
14. |
D5S818 |
10, 11 |
10, 11 |
2.6947 |
DYS437 |
15 |
15 |
|
15. |
FGA |
22, 27 |
26, 27 |
50 |
DYS438 |
9 |
9 |
|
16. |
Amelogenin |
X |
X |
CPI = 257341.55 |
DYS448 |
18 |
18 |
Table: 1 Autosomal-STR and Y-STR DNA profiles of alleged father and child generated using AmpFLSTRTM Identifiler PlusTM PCR amplification kit and AmpFLSTRTM YfilerTM PCR Amplification Kit (Applied Biosystems) respectively and calculation of Combined Paternity Index (CPI).
Establishment Of Paternity Through Statistical Analysis
The paternity index was calculated as PI - x/y , where X is the likelihood ratio of alleged father being the biological father (null hypothesis), and Y is the probability (population allele frequency) of a random man from the population (alternative hypothesis for all other possible outcomes) [25]. The value of Y (autosomal STR frequency) was obtained from the population STR frequency data of Jharkhand State published by the State Forensic Science Laboratory Jharkhand, India [26]. The Combined Paternity Index (CPI) was obtained by multiplying all individual Paternity Indices (PI) calculated for each locus. CPI is the measure of the strength of genetic evidence that tries to fit in the hypothesis that the alleged man is the father of the child as compared to someone else from the population. The calculated CPI was 257341.55 (Table 1) which is far greater than the critical value of 1. To test the strength of the hypothesis again that the tested man is the biological father rather than a random man from the population based on non-genetic evidence, the Prior Probability of Paternity (POP) was calculated as POP = (CPI/CPI+1)= 0.999997396 which gave result close to the certainty value of 1, where value of 0.5 is a blind assumption by the court system from the testimony of father, mother, and other witnesses, meaning equal chances of both hypotheses to occur.
Results and Discussions
In this case, autosomal and Y chromosomal STR profiles (Supplementary Figures S1-S4) were generated from standard reference blood samples. The paternity was established using paternal allelic inclusion of the child in the autosomal DNA profile of alleged father and statistical analysis mentioned above (Table 1). The diallelic STR marker DYS385 was found to be monoallelic, a very unusual observance, while two short arm markers (Amelogenin and DYS458) were not amplified, indicating Y chromosome micro-deletion. The frequency of deletion incidence of both DYS458 and AMEL Y loci in the human Y chromosome is very rare, a very common pattern of microdeletion which is observed across the world mostly among the Asian and North American population [27,28]. This is the first case of paternity determined and reported across globe, where noted Y chromosomal allelic abnormalities (Y chromosomal microdeletion along with DYS385 monoallelism) in both the father and his biological male child (Table 1), have no effect on the virility/fertility of human beings.
Conflict of Interest
The authors certify that there is no academic or financial interest accrued to anyone and no other conflicts of interest exists whatsoever.
Authors' Contributions
AKR: Supervision, Data curation, Writing- Original draft preparation.
RR: Performed Experiment. All authors have read and approved the final manuscript. All authors confirm the authenticity of all raw data.
Funding
No funding source is available.
References
- Kayser M (2017) Forensic use of Y-chromosome DNA: A general overview. Hum Genet 136: 621-35.
- Sullivan KM, Mannucci A, Kimpton CP, Gill P (1993) A rapid and quantitative DNA sex test: Fluorescence-based PCR analysis of X-Y homologous gene amelogenin. Biotechniques 15: 636-638, 640-641.
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, et al. (1990) Genetic evidence equating SRY and the testis-determining factor. Nature 348: 448-450.
- Colaco S, Modi D (2018) Genetics of the human Y chromosome and its association with male infertility. Reprod Biol Endocrinol 16: 14.
- Brown KE, Barnett MA, Burgtorf C, Shaw P, Buckle VJ, et al. (1194) Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum Mol Genet 3: 1227-1237.
- Helena Mangs A, Morris BJ (2007) The Human Pseudoautosomal Region (PAR): Origin, Function and Future. Curr Genomics 8: 129-136.
- Dupuy O, Palou M, Mayaudon H, Sarret D, Bordier L, et al. (2001) [De La Chapelle syndrome]. Presse Med 30: 369-372.
- Lashkari FM, Totonchi M, Zamanian MR, Mansouri Z, Sadighi Gilani MA, et al. (2017) 46,XX males: A case series based on clinical and genetics evaluation. Andrologia 49.
- Jehan Z, Vallinayagam S, Tiwari S, Pradhan S, Singh L, et al. (2007) Novel noncoding RNA from human Y distal heterochromatic block (Yq12) generates testis-specific chimeric CDC2L2. Genome Res 17: 433-440.
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, et al. (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825-837.
- Bachtrog D (2013) Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14: 113-124.
- Wilson J, Staley JM, Wyckoff GJ (2020) Extinction of chromosomes due to specialization is a universal occurrence. Sci Rep10: 2170.
- Balanovsky O (2017) Toward a consensus on SNP and STR mutation rates on the human Y-chromosome. Hum Genet 136: 575-590.
- Ballantyne KN, Goedbloed M, Fang R, Schaap O, Lao O, et al. (2010) Mutability of Y-chromosomal microsatellites: Rates, characteristics, molecular bases, and forensic implications. Am J Hum Genet 87: 341-353.
- Boattini A, Sarno S, Mazzarisi AM, Viroli C, De Fanti S, et al. (2019) Estimating Y-Str Mutation Rates and Tmrca Through Deep-Rooting Italian Pedigrees. Sci Rep 9: 9032.
- Akinsal EC, Baydilli N, Dundar M, Ekmekcioglu O (2018) The frequencies of Y chromosome microdeletions in infertile males. Turk J Urol 44: 389-392.
- Spies M, Fishel R (2015) Mismatch repair during homologous and homeologous recombination. Cold Spring Harb Perspect Biol 7: a022657.
- Dumanski JP, Lambert JC, Rasi C, Giedraitis V, Davies H, et al. (2016) Mosaic Loss of Chromosome Y in Blood Is Associated with Alzheimer Disease. Am J Hum Genet 98: 1208-1219.
- Forsberg LA (2017) Loss of chromosome Y (LOY) in blood cells is associated with increased risk for disease and mortality in aging men. Hum Genet 136: 657-663.
- Burki TK (2014) Smoking and mosaic Y chromosome loss. Lancet Oncol 16: e12.
- Dumanski JP, Rasi C, Lonn M, Davies H, Ingelsson M, et al. (2015) Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science 347: 81-83.
- Barbisin M, Fang R, O'Shea CE, Calandro LM (2009) Developmental validation of the Quantifiler Duo DNA Quantification kit for simultaneous quantification of total human and human male DNA and detection of PCR inhibitors in biological samples. J Forensic Sci 54: 305-319.
- Wang DY, Chang CW, Lagace RE, Calandro LM, Hennessy LK (2011) Developmental validation of the AmpFlSTR(R) Identifiler(R) Plus PCR Amplification Kit: An established multiplex assay with improved performance. J Forensic Sci 57: 453-465.
- Mulero JJ, Chang CW, Calandro LM, Green RL, Li Y, et al. (2006) Development and validation of the AmpFlSTR Yfiler PCR amplification kit: A male specific, single amplification 17 Y-STR multiplex system. J Forensic Sci 51: 64-75.
- Gjertson DW, Brenner CH, Baur MP, Carracedo A, Guidet F, et al. (2007) ISFG: Recommendations on biostatistics in paternity testing. Forensic Sci Int Genet 1: 223-231.
- Imam J, Reyaz R, Singh RS, Bapuly AK, Shrivastava P (2018) Genomic portrait of population of Jharkhand, India, drawn with 15 autosomal STRs and 17 Y-STRs. Int J Legal Med 132: 139-140.
- Chang YM, Perumal R, Keat PY, Yong RY, Kuehn DL, et al. (2007) A distinct Y-STR haplotype for Amelogenin negative males characterized by a large Y(p)11.2 (DYS458-MSY1-AMEL-Y) deletion. Forensic Sci Int 166: 115-120.
- Borovko S, Shyla A, Korban V, Borovko A (2014) Amelogenin test abnormalities revealed in Belarusian population during forensic DNA analysis. Forensic Sci Int Genet 15: 98-104.
Supplementary Figures S1-S4
The autosomal profile of alleged father – Figure S1 and autosomal profile of male child – Figure S2 were generated using AmpFLSTRTM Identifiler PlusTM PCR Amplification Kit (Applied Biosystems) while Y-STR profile of alleged father – Figure S3 and Y-STR of male child – Figure S4 were generated using AmpFLSTRTM YfilerTM PCR Amplification Kit (Applied Biosystems).
Ethics approval and consent to participate
The blood of each participant was taken in the presence of two witnesses and certified in Blood Authentication Form as per the rules of Jharkhand Police Manual. The blood samples were collected by an authorized and certified medical officer on sterile gauze cloth pieces from the alleged father and child as per direction from the court of law (The District and First Additional Session Judge cum Special Judge, Latehar) and sent to the State Forensic Science Laboratory Jharkhand for DNA examination. This case imposes the Section 376(3) Indian Penal Code (IPC) Act and Section 4(2) of Protection of Children from Sexual Offences (POCSO) Act, and hence consent is not required for such heinous crime cases involving imprisonment of more than seven years.
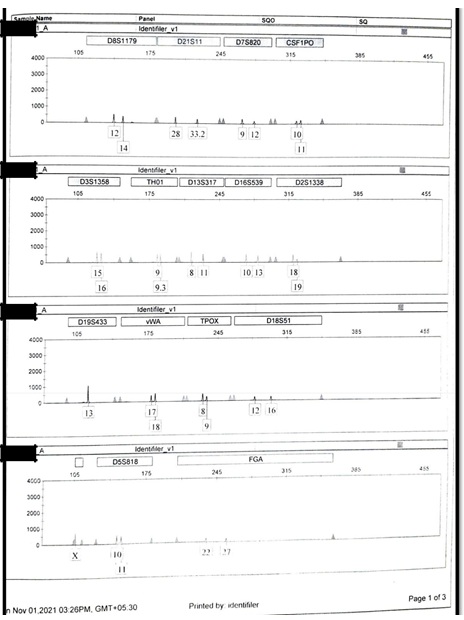 Figure S1.
Figure S1.
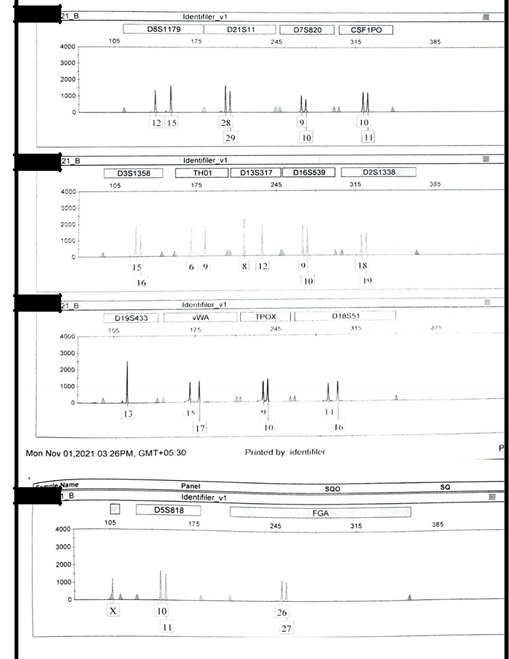 Figure S2.
Figure S2.
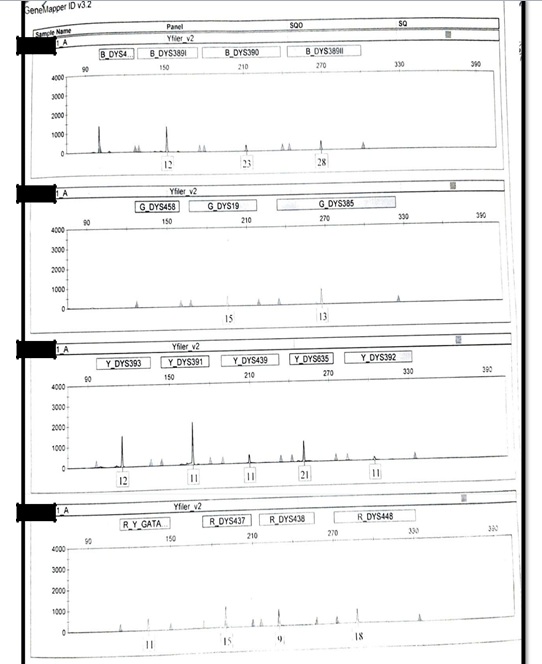 Figure S3.
Figure S3.
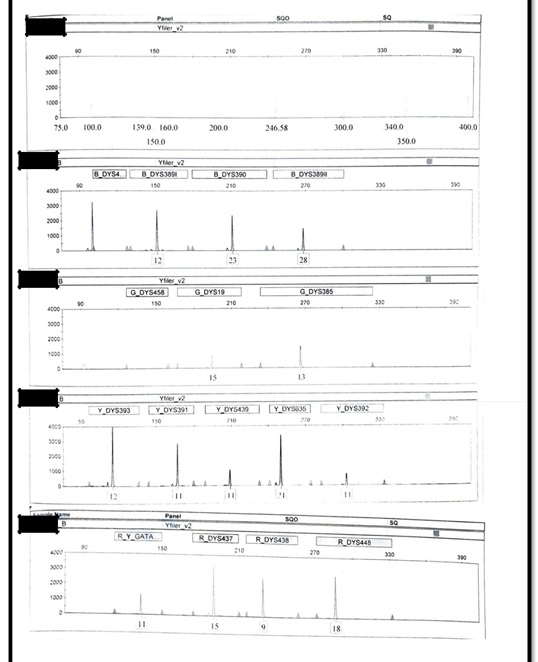 Figure S4.
Figure S4.
Citation: Rana AK, Reyaz R (2022) A Rare Case of Dys385 Monoallelism Along With Amel Y Deletion: A Case Report. Forensic Leg Investig Sci 8: 066.
Copyright: © 2022 Ajay Kumar Rana, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

