
Collection Methods for Touch DNA Direct Amplification
*Corresponding Author(s):
Alketbi Salem KUniversity Of Central Lancashire, Preston, United Kingdom
Tel:00447774141205,
Email:alkitbe.11@hotmail.com
Abstract
DNA direct amplification from biological samples such as saliva and blood has proven to be successful but can be more challenging from minute samples such as trace DNA. Developing new protocols to collect and process Touch DNA samples can improve DNA direct amplification, therefore a variety of collection methods were investigated for their efficiency to collect Touch DNA for direct amplification using a GlobalFiler™ PCR amplification Kit. Touch Samples were collected from six surfaces to test the sensitivity of the collection method for direct amplification, with the number of alleles observed from the DNA profiles significantly affected by collection type (p < 0.001). Copan microFLOQ swabs (MF) and SceneSafe Fast™ Minitapes (MT) recovered a higher percentage of alleles and were more reliable for direct DNA amplification.
Keywords
Cotton swab; DNA direct amplification; DNA recovery; Forensic science; GlobalFiler™ PCR amplification kit; MicroFLOQ™ swabs; Nylon flocked swab; SceneSafe fast™ minitape; Touch DNA; Trace DNA
Introduction
Touch DNA is present on many used items but is often challenging [1-13]. However, direct amplification in DNA profiling omits the DNA extraction and quantification steps, directly amplifying the DNA by PCR after sample collection. This technique maximises the quantity of DNA, avoids laboratory personnel errors, minimises DNA contamination from handling, as well as reduces the overall DNA sample processing time and cost [1]. While DNA direct amplification from biological samples such as saliva and blood is successful [14], it can be more challenging from minute samples such as trace DNA [1,15]. Developing new protocols to collect and process Touch DNA samples can improve DNA direct amplification, therefore a variety of collection methods were tested for their efficiency to collect Touch DNA for direct amplification using a GlobalFiler™ PCR amplification Kit.
Materials And Methods
Experimental setup
The deposition methodology was as previously described [3]. Briefly, two participants previously identified as high and low shedders were asked to clean their hands with antibacterial soap and refrain from doing any activity for 10 min, then charge the fingers of both hands with eccrine sweat by touching their forehead to load them with epithelial cells. The participants were then instructed to touch the surfaces using their index, middle, and ring fingers of both hands separately for deposition by applying medium pressure on a 2.5 x 3.5 cm area for 1 min (total number of deposits is 72, 12 replicates per each collection method). Touch DNA was deposited on a variety of surfaces including stainless steel, textured plastic, textured wood, copier paper, fabric (composed of 65% polyester and 35% cotton), and glass. All non-porous surfaces were sterilised with 2% virkon (viricidal disinfectant) and Ultra Violet radiation (UV) for 20 min, and porous surfaces were irradiated using UV for 30 min.
DNA recovery
DNA was collected using six collection methods as shown in (Figure 1). The MicroFLOQ™ swab (MF), Mini Cotton Swab (MCS) and Mini Plastic Swab (MPS) were moistened with 1μL of sterile distilled water using a pipette. The Cotton Swab (CS) was moistened with 100μL of sterile distilled water applied using a plastic spray bottle technique [7] and the nylon flocked Swab (NS) was moistened with 30μL of sterile distilled applied by pipette. No water was added to Minitapes (MT) but to increase the amount of Touch DNA collected, each minitape was applied 16 times to the area.
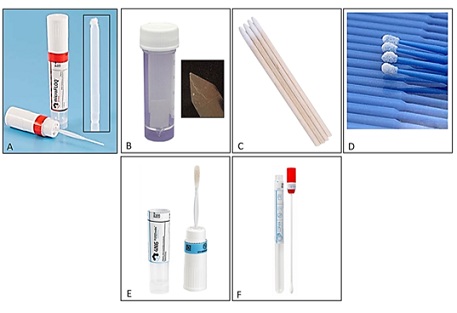 Figure 1: Six collection methods used for direct amplification: (A) MicroFLOQ™ swab (MF) (Copan), (B) SceneSafe Fast™ Minitape (MT) (K545), (C) Mini Cotton Swab (MCS) (Fenshine), (D) Mini Plastic Swab (MPS) (G2Plus), (E) Copan Nylon flocked Swab (4N6FLOQSwabs®) (NS) and (F) Copan Cotton Swab (CS) (150C).
Figure 1: Six collection methods used for direct amplification: (A) MicroFLOQ™ swab (MF) (Copan), (B) SceneSafe Fast™ Minitape (MT) (K545), (C) Mini Cotton Swab (MCS) (Fenshine), (D) Mini Plastic Swab (MPS) (G2Plus), (E) Copan Nylon flocked Swab (4N6FLOQSwabs®) (NS) and (F) Copan Cotton Swab (CS) (150C).
DNA amplification and analysis
The DNA samples were transferred to PCR tubes (0.2 ml) for direct amplification using the GlobalFiler™ PCR Amplification Kit (Thermo Fisher Scientific). The MF swab head was broken directly into the tube and approximately 2 mm2 of the other swabs and MT were cut and placed into the tube. The PCR master mix was added directly to the tubes and the reaction volume was made up with TE buffer before amplification was performed on an ABI GeneAmp® 9700 PCR System (Life Technologies) for 29 cycles. The amplified products were size-separated and detected on an ABI 3500 Genetic Analyzer (Life Technologies) using 1 μl of PCR product, 9.6 μl Hi-Di™ formamide, and 0.4μl GeneScan™ 600 LIZ® Size Standard v2.0 (Thermo Fisher Scientific) according to the manufacturer’s protocol. Statistical analysis was performed with RStudio using factorial analysis of variance (ANOVA) and Microsoft Excel.
Results And Discussion
The number of alleles observed in the DNA profiles was significantly affected by collection type (p < 0.001), as well as surface type (p < 0.05) (Figure 2 & 3). MicroFLOQ swabs (MF) and Minitapes (MT) recovered a higher percentage of alleles and were more reliable for direct DNA amplification than Mini Plastic Swabs (MPS) and Mini Cotton Swabs (MCS) (p < 0.05). Cotton Swabs (CS) and 4N6FLOQSwabs (NS) recovered a moderate percentage of alleles but the results were not consistent compared to MF and MT.
 Figure 2: Percentage of alleles recovered from Touch DNA samples (n= 72) collected by six different methods and directly amplified using the GlobalFiler™ kit. MF; microFLOQ™ swabs - MPS; Mini Plastic Swabs - MCS; Mini Cotton Swabs - MT; Minitapes - CS; Cotton Swabs - NS; Nylon flocked Swabs.
Figure 2: Percentage of alleles recovered from Touch DNA samples (n= 72) collected by six different methods and directly amplified using the GlobalFiler™ kit. MF; microFLOQ™ swabs - MPS; Mini Plastic Swabs - MCS; Mini Cotton Swabs - MT; Minitapes - CS; Cotton Swabs - NS; Nylon flocked Swabs.
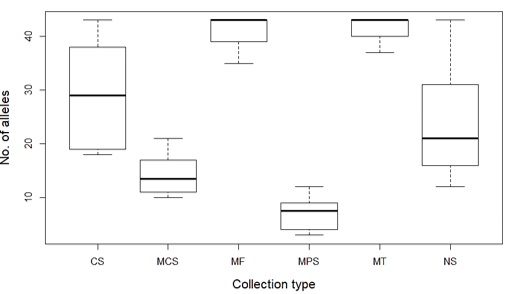 Figure 3: Number of alleles recovered from Touch DNA samples (n= 72) collected by six different methods and directly amplified with the GlobalFiler™ kit. CS; Cotton Swabs - MCS; Mini Cotton Swabs - MF; microFLOQ™ swabs - MPS; Mini Plastic Swabs - MT; Minitapes - NS; Nylon flocked Swabs.
Figure 3: Number of alleles recovered from Touch DNA samples (n= 72) collected by six different methods and directly amplified with the GlobalFiler™ kit. CS; Cotton Swabs - MCS; Mini Cotton Swabs - MF; microFLOQ™ swabs - MPS; Mini Plastic Swabs - MT; Minitapes - NS; Nylon flocked Swabs.
More alleles were recovered from non-porous surfaces than porous surfaces (p < 0.05), when samples were processed for direct PCR (allele recovery for Stainless steel; 77%, Glass; 72%, Textured plastic; 65%, Textured wood; 56%, Copier paper; 49%, and Fabric; 51%). Touch DNA samples were collected from six surfaces to test the sensitivity of the collection method for direct amplification as it has been shown previously that different surface types impact the recovery of Touch DNA [3]. MF and MT yielded full and partial STR profiles with balanced loci but the other collection methods tested only produced partial STR profiles or no data (Figure 4). Direct amplification resulted in artifacts in some samples, mostly split and shoulder peaks (Figure 5).
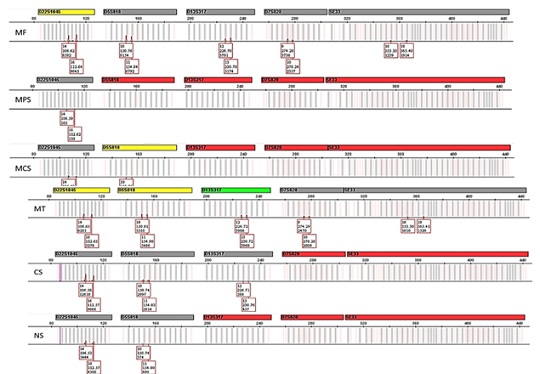 Figure 4: Comparison of electropherograms at five loci (D22S1045, D5S818, D13S317, D7S820 and SE33) generated by the direct amplification of samples collected by the six different methods. MF; microFLOQ™ swabs - MPS; Mini Plastic Swabs - MCS; Mini Cotton Swabs - MT; Minitapes - CS; Cotton Swabs - NS; Nylon flocked Swabs.
Figure 4: Comparison of electropherograms at five loci (D22S1045, D5S818, D13S317, D7S820 and SE33) generated by the direct amplification of samples collected by the six different methods. MF; microFLOQ™ swabs - MPS; Mini Plastic Swabs - MCS; Mini Cotton Swabs - MT; Minitapes - CS; Cotton Swabs - NS; Nylon flocked Swabs.
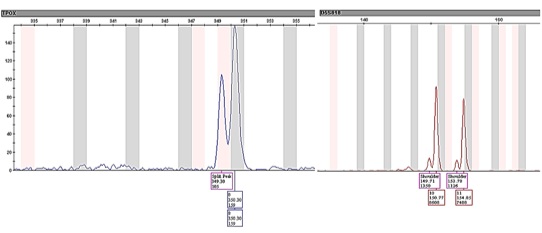 Figure 5: Split and shoulder peaks present in the STR profiles produced by the direct amplification of DNA samples collected by the six different methods.
Figure 5: Split and shoulder peaks present in the STR profiles produced by the direct amplification of DNA samples collected by the six different methods.
Conclusion
The success of the direct amplification of Touch DNA is dependent on the quantity of cellular material available on the surface but can be improved by an effective collection method. MicroFLOQ swabs and Minitapes were the most effective methods tested in this study to collect trace DNA for direct amplification. The MicroFLOQ swabs with their appropriate design allow easy collection and sample processing compared to the Minitapes which require more handling.
Conflict of Interest
None
Acknowledgements
This study was approved by the General Department of Forensic Science and Criminology in Dubai Police and ethical approval was granted by the University of Central Lancashire’s Research Ethics Committee (ref. no. STEMH 912).
References
- Alketbi SK (2018) The affecting factors of Touch DNA. Journal of Forensic Research 9: 424.
- Alketbi SK (2020) Collection of Touch DNA from rotten banana skin. International Journal of Forensic Sciences 5: 000204.
- Alketbi SK, Goodwin W (2019) The effect of surface type, collection, and extraction methods on Touch DNA. Forensic Science International Genetics Supplement Series 7: 704-706.
- Alketbi SK, Goodwin W (2019) The effect of time and environmental conditions on Touch DNA. Forensic Science International Genetics Supplement Series 7: 701-703.
- Alketbi SK, Goodwin W (2019) The effect of sandy surfaces on Touch DNA. Journal of Forensic Legal & Investigative Sciences 5: 034.
- Alketbi SK, Goodwin W (2021) Touch DNA Collection Techniques for Non-Porous Surfaces Using Cotton and Nylon Swabs. Journal of Scientific & Technical Research 36: 28608-28612.
- Alketbi SK, Goodwin W (2019) Validating Touch DNA collection techniques using cotton swabs. Journal of Forensic Research 10: 445.
- Alketbi SK (2022) An Innovative Solution to Collect Touch DNA for Direct Amplification. Journal of forensic sciences & criminal investigation 16: 555928.
- Alketbi SK, Alsoofi S (2022) Dual Recovery of DNA and Fingerprints using Minitapes. Journal of forensic sciences & criminal investigation 16: 555929.
- Alketbi SK, Goodwin W (2022) The Impact of Area Size and Fabric Type on Touch DNA Collected from Fabric. Journal of forensic sciences & criminal investigation 16: 555926.
- Alketbi SK (2022) The Impact of Collection Method on Touch DNA Collected from Fabric. Journal of forensic sciences & criminal investigation 15: 555922.
- Alketbi SK, Goodwin W (2022) The impact of deposition area and time on Touch DNA collected from fabric. Forensic Science International. Genetics Supplement Series 8: 45-47.
- Alketbi SK (2022) Analysis of Touch DNA [Unpublished doctoral dissertation]. University of Central Lancashire.
- Ambers A, Wiley R, Novroski N, Budowle B (2018) Direct PCR amplification of DNA from human blood stains, saliva, and touch samples collected with microFLOQ® Forensic Science International Genetics 32: 80-87.
- Linacre A, Pekarek V, Swaran YC, Tobe SS (2009) Generation of DNA profiles from fabrics without DNA extraction. Forensic Science International Genetics 4: 137-141.
Citation: Salem SK, Goodwin W (2023) Collection Methods for Touch DNA Direct Amplification. J Forensic Leg Investig Sci 9: 072.
Copyright: © 2023 Alketbi Salem K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

